Abstract
Aims/hypothesis
Adult beta cells in the pancreas are the sole source of insulin in the body. Beta cell loss or increased demand for insulin impose metabolic challenges because adult beta cells are generally quiescent and infrequently re-enter the cell division cycle. The aim of this study is to test the hypothesis that a family of proto-oncogene microRNAs that includes miR-17-92 and miR-106b-25 clusters regulates beta cell proliferation or function in the adult endocrine pancreas.
Methods
To elucidate the role of miR-17-92 and miR-106b-25 clusters in beta cells, we used a conditional miR-17-92/miR-106b-25 knockout mouse model. We employed metabolic assays in vivo and ex vivo, together with advanced microscopy of pancreatic sections, bioinformatics, mass spectrometry and next generation sequencing, to examine potential targets of miR-17-92/miR-106b-25, by which they might regulate beta cell proliferation and function.
Results
We demonstrate that miR-17-92/miR-106b-25 regulate the adult beta cell mitotic checkpoint and that miR-17-92/miR-106b-25 deficiency results in reduction in beta cell mass in vivo. Furthermore, we reveal a critical role for miR-17-92/miR-106b-25 in glucose homeostasis and in controlling insulin secretion. We identify protein kinase A as a new relevant molecular pathway downstream of miR-17-92/miR-106b-25 in control of adult beta cell division and glucose homeostasis.
Conclusions/interpretation
The study contributes to the understanding of proto-oncogene miRNAs in the normal, untransformed endocrine pancreas and illustrates new genetic means for regulation of beta cell mitosis and function by non-coding RNAs.
Data availability
Sequencing data that support the findings of this study have been deposited in GEO with the accession code GSE126516.
Similar content being viewed by others
Introduction

MicroRNAs (miRNAs) are small, non-coding RNAs that provide a broad post-transcriptional silencing mechanism [1], including in metabolism and diabetes [2,3,4]. miRNAs are essential for normal beta cell function and inactivation of miRNA biogenesis in beta cells results in a diabetic phenotype [5, 6].
Beta cell mass is a function of cell number and size, correlates with body demand [7] and is controlled by beta cell replication [8,9,10]. Cell division is a tightly regulated process, with four main stages, and three checkpoints (i.e. G1/S, G2/M and mitotic checkpoint) that guarantee the accomplishment of necessary molecular activities before progression to the next stage [11]. Aberrant cell cycle progression might result in cell cycle failure, premature cell cycle exit and cell death. In beta cells, impaired proliferation may result in low insulin levels and hyperglycaemia/diabetes; therefore better understanding of the molecular mechanisms controlling beta cell proliferation is valuable.
The miR-17-92 family contains 15 miRNAs that regulate cell proliferation and apoptosis [12,13,14]. These miRNAs are transcribed from three polycistronic clusters (miR-17-92 on mouse chromosome 13, miR-106a-363 on chromosome X and miR-106b-25 on chromosome 5). The clusters share four main ‘seed’ subtypes and hence joint downstream mRNA targets [15]. Genetic deletion of miR-17-92 results in smaller mouse embryos with severely hypoplastic lungs. Furthermore, deletion of both miR-17-92 and the homologous miR-106b-25 cluster is lethal for embryos [13].
Expression of miR-17-92 family members is regulated during the cell cycle, at least in cultured cells, with the highest levels measured at the G2/M transition [16]. This may allow inhibition of target proteins involved in the transition between the G1/S phases. Indeed, miR-17-92 family members are interwoven into a regulatory network, wherein expression of these miRNAs is induced by c-Myc and E2F and the miRNAs repress the expression of E2F family members through conserved binding sites at the 3′UTR of E2F1/2/3 [17,18,19].
The Regazzi laboratory demonstrated roles for the miR-17-92 family in metabolic adaptation of beta cells in newborn rats to changes in nutrient supply [20] and in regulating islet circadian gene expression [21]. We hypothesised that miR-17-92/miR-106b-25 family members regulate adult beta cell division, given the interaction of this miRNA family with c-Myc, a known driver of beta cell proliferation [22].
Methods
Mouse strains
Female and male c57bl/6 mice were housed and handled at the Weizmann Institute of Science and in accordance with protocols approved by the Institutional Animal Care and Use Committee of the Weizmann Institute of Science. All mice we used were bred in-house. To generate Pdx1-Cre;miR-17-92LoxP/LoxP;miR-106-25−/− (miR-17-92/miR-106b-25-KO) mice, we previously crossed Pdx1-Cre transgenic mice [23] (a gift from D. Melton [Howard Hughes Medical Institute, Harvard University, Boston, MA, USA]) with miR-17-92LoxP/LoxP and further with miR-106b-25−/− mice [13] (both were gifts from T. Jacks [Howard Hughes Medical Institute, Massachusetts Institute of Technology, Boston, MA, USA] and A. Ventura [Memorial Sloan Kettering Cancer Center, New York, NY, USA]).
ROSA-miR-17-92conditional overexpressing mice (were a gift from K. Rajewsky [Max Delbrück Center for Molecular Medicine, Berlin, Germany]) [24] and were crossed with Pdx1-Cre to achieve Pdx1-Cre;ROSA-miR-17-92conditional mice. CcnB1-GFP transgenic mice were generated by Y. Dor (The Hebrew University of Jerusalem, Israel) [25, 26].
Isolation of islets of Langerhans, flow cytometry and cell sorting
Islets were isolated using collagenase P (Roche, Switzerland) injected into the pancreatic duct, followed by Histopaque gradient (1119, 1083 and 1077; Sigma-Aldrich, Israel) as described in [27]. For miRNA profiling along the beta cell cycle, flow cytometry, islet dissociation and cell sorting were performed as described in [25].
Pancreas physiology assays
Blood glucose was determined using an Ascensia elite glucometer (Ascensia, Switzerland). Insulin levels in the pancreas and serum were determined using an ultrasensitive insulin ELISA kit (90,080; Crystal Chem, Elk Grove Village, IL, USA). GTTs and glucose-stimulated insulin secretion (GSIS) tests were performed by injecting glucose (2 mg/g) intraperitoneally after mice were fasted overnight (~18 h) at different time points (age 4 weeks to 12 months). ITT was performed by injecting insulin (0.8 U/g) intraperitoneally after 4- to 6-week-old mice were fasted for 5 h. Blood for GTT and ITT was repeatedly sampled from the tail vein. Retro-orbital blood was sampled before the injection and 15 min post-injection for the in vivo GSIS test. Insulin secreted to the medium in the ex vivo GSIS was measured after 1 h of incubation with either 2.5 or 25 mmol/l glucose by fluorescence resonance energy transfer (FRET) (62IN2PEG; Cisbio, France). Protein kinase A (PKA) activity was quantified using a PKA Kinase Activity Assay Kit (ab139435; Abcam, UK). Islets for all in vitro assays were purified from 4- to 6-week-old mice.
Static and dynamic stimulation of insulin secretion
Insulin secretion studies were performed in KRB containing 114.4 mmol/l NaCl, 5 mmol/l KCl, 24 mmol/l NaHCO3, 1 mmol/l MgCl2, 2.2 mmol/l CaCl2, 10 mmol/l HEPES and 0.5% wt/vol. BSA, adjusted to pH 7.35. In static incubation experiments, 10–20 islets from 4- to 6-week-old mice were pre-incubated in basal KRB containing 2.5 mmol/l glucose for 1 h. Islets were consecutively incubated at 2.5 and 25 mmol/l glucose for 1 h each. Medium was collected at the end of each incubation period. Insulin assays were performed in Eppendorf tubes at 37°C and 5% CO2.
A perifusion system (Biorep, Miami Lakes, FL, USA) equipped with a peristaltic pump was used for dynamic assessment of insulin secretion. Forty size-matched islets were placed in columns and perifused at a flow rate of 100 μl/min with KRB (basal glucose concentration 2.8 mmol/l) at 37°C. After equilibration, high glucose (16.7 mmol/l) KRB was used. Insulin secreted to the medium was collected in 96-well plates, quantified by FRET (Cisbio) or ELISA (Crystal Chem) and normalised to total islet insulin content.
Pancreatic histology and immunohistochemistry
Pancreases from 4- to 6-week-old mice were dissected and fixed in 4% vol./vol. paraformaldehyde for 24 h at 4°C and then processed into paraffin blocks. Sections (5 μm thick) were de-paraffinised, rehydrated and antigen retrieval was performed using a PickCell pressure cooker (PickCell, the Netherlands). The following primary antibodies were used: guinea pig anti-insulin (1:200, A05641; Dako, Denmark); rabbit anti-activated caspase-3 (1:50, c-96,615; Cell Signaling, Danvers, MA, USA); rabbit anti-Ki67 (1:200, SP6; Cell Marque, Rocklin, CA, USA), mouse anti-bromodeoxyuridine (BrdU) (1:200, RPN202; GE Healthcare, Chicago, IL, USA) and phosphorylated histone H3 (PHH3) (1:200, c-9701; Cell Signaling). For TUNEL staining we used the ApopTag red in situ apoptosis detection kit (s7165; Merck, Germany). For DNA counter-stain, we used Hoechst 33342 (1 μg/ml, H3570; Thermo Fisher, Waltham, MA, USA). Secondary antibodies conjugated to CY2, CY3 or CY5 were all from Jackson Immunoresearch Laboratories Baltimore, MD, USA (1:200). All the antibodies were previously validated, and all immunostaining included a negative control (no primary antibody); a positive control was also used for the apoptosis staining. All primary and secondary antibodies were diluted in CAS-block (008120 Thermo Fisher). Fluorescence images were captured using a Zeiss LSM710/780/800 Laser Scanning/confocal microscope system equipped with a Zeiss camera with ×40 / ×63 magnification (Thornwood, NY, USA).
Histomorphometry
Digital images of consecutive paraffin-embedded pancreas sections (50 μm apart, spanning the entire pancreas, approximately 40 sections/pancreas) were obtained at a low magnification (×20) and stitched using NIS-Elements software (Nikon, Japan) and 3DHistech (Hungary) Pannoramic Viewer. The fraction of insulin-positive surface was determined by insulin immunoreactivity and the whole pancreas area was determined by haematoxylin counter-stain. Beta cell mass was calculated as the product of pancreas weight and the fraction of tissue covered by beta cells.
RNA quantification
Extraction of total RNA was carried out by the miRNeasy Mini Kit (Qiagen, Germany). mRNA cDNA was synthesised using an oligo d(T) primer (C1101; Promega, Madison, WI, USA) and SuperScript II reverse transcriptase (18064-014; Invitrogen, Carlsbad, CA, USA). Synthesis of miRNA cDNA was created using Taqman MicroRNA qPCR Assays (Applied Biosystems, Foster City, CA, USA). mRNA quantitative real-time RT-PCR (qRT-PCR) analysis was performed on a LightCycler 480 System (Roche) using Kapa SYBR Green qPCR kit (Finnzymes, Finland). miRNA qRT-PCR was performed on ABI Step one (Thermo Fisher). miRNA and mRNA levels were normalised to the expression of small RNAs (sno234 and U6) or mRNA (Gapdh and Hprt), respectively.
Mouse embryonic fibroblast isolation and adenovirus infection
miR-17-92/miR-106b-knockout (KO) or control mouse embryonic fibroblasts (MEFs) were harvested as in [28], plated at 50–60% confluency and grown in monolayer cultures in DMEM supplemented with 20% vol./vol. FBS (Biological Industries, Israel), 1% vol./vol. penicillin–streptomycin, 1% vol./vol. l-glutamine, 1% vol./vol. sodium pyruvate and 1% vol./vol. MEM-non-essential amino acid (Biological Industries). Cells were infected the next day with Ad5CMVeGFP (eGFP-adenovirus) or Ad5CMVCRE-eGFP (CRE-adenovirus), 300 viral particles/cell (Gene Transfer Vector Core, University of Iowa). Medium was added after 24 h, replaced after 48 h and cells were harvested 5 days post-infection.
RNA sequencing
cDNAs were sequenced on Illumina 2500 (Ilumina, San Diego, CA USA) sequencing machine with 50 bp single read protocol. Reads for each sample were mapped independently using TopHat2 version (https://ccb.jhu.edu/software/tophat/index.shtml) (v2.0.10) [29] against the mouse genome build mm9. Approximately, 85–90% mapping rate was observed. Only uniquely mapped reads were used to determine the number of reads falling into each gene with the HTSeq-count script (https://htseq.readthedocs.io/en/release_0.11.1/count.html) (0.6.1p1) [30]. Differentially expressed genes were calculated with the DESeq2 package (v1.4.5) [31]. Genes that were expressed on at least one sample were considered. Differentially expressed genes were determined by p value <0.05 and an absolute fold change >1.5. Benjamini–Hochberg correction was used to adjust p value with false discovery rate (FDR) <0.05. Hierarchical clustering using Pearson dissimilarity and complete linkage was performed in order to explore a pattern of gene expression. Clustering analysis was performed with Matlab software (https://www.mathworks.com/products/matlab.html) (8.0.0.783). Gene ontology (GO) term enrichment analysis was performed using DAVID (https://david.ncifcrf.gov/) [32, 33].
Sequencing data that support the findings of this study have been deposited in GEO with the accession codes GSE126516 [34].
Mass spectrometry
The samples were subjected to in-solution tryptic digestion followed by a desalting step. The resulting peptides were analysed using nanoflow liquid chromatography (nanoAcquity, Milford, MA, USA) coupled to high-resolution, high-mass-accuracy MS (Q Exactive Plus, Thermo Fisher). Samples were separately analysed in random order. Data were normalised to the sample total ion current and searched against the mouse protein database, to which a list of common laboratory contaminants was added (Mascot algorithm). Quantitative analysis was performed using Genedata Expressionist (UK). Only proteins identified by more than two peptides and more than nine amino acids/peptides were considered. p values were corrected for multiple hypothesis using Benjamini–Hochberg procedure with FDR <0.05. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [35] partner repository with the dataset identifier PXD012610.
Xist single molecule fluorescence in situ hybridisation and image analysis
Single molecule in situ hybridisation (smFISH) was as in [36]. TransQuant (https://ars.els-cdn.com/content/image/1-s2.0-S1046202315301559-mmc1.zip; accessed 13 Dec 2018) was used for Xist smFISH signal segmentation and analysis [37]. Ilastik (https://www.ilastik.org/) (1.3.1) was used for cell cycle image segmentation [38].
Statistical analysis
Data are expressed as means (SEM) and a two-sided Student’s t test was used for statistical comparisons.
Results
miR-17-92 expression in developing and adult mouse endocrine pancreas
miR-17 and miR-20a are expressed only from the miR-17-92 cluster, miR-363 from miR-106a-363 and miR-25 from miR-106b-25 cluster (Fig. 1a), enabling the discrimination of expressed clusters by qRT-PCR. miR-106a-363 expression was undetected, consistent with its reported limited expression pattern [13], whereas miR-17-92 and miR106b-25 clusters were expressed in mouse embryonic pancreas (Fig. 1b).
Dynamics of miRNA expression from the three paralogous genomic clusters encoding the miR-17-92 family members in the developing pancreas of the mouse. (a) Diagram of the genomic organisation of the miR-17-92 and miR-106b-25 clusters. miRNAs are colour coded according to seed sequences that define target specificity. (b) Relative expression levels of miR-17, miR-25 (miR-106b-25 cluster, chromosome 5), miR-20a (miR-17-92 cluster, chromosome 13) and miR-363 (miR-106a-363 cluster, chromosome X) in pancreatic buds, isolated from mouse embryos at embryonic day E14.5. Expression relative to miR-17 is shown and is normalised to the expression of the non-coding RNA U6. It is noteworthy that miR-363 is undetected. n=3. (c) miR-17, miR-20a and miR-25 expression at multiple time points in the pancreas during embryonic development. miRNA expression is normalised to U6 and to expression at E12.5. RNA for the embryonic measurements was extracted from three embryonic pancreatic buds (three embryos) for each sample. (d, e) mRNA levels of Ki67 and Top2a, two cell cycle markers (d), and of specific miRNAs (e), in sorted replicating vs non-replicating beta cells. Data are shown as mean ± SEM and expression is relative to that in non-replicating beta cells. *p<0.05 and ***p<0.001 (two-sided Student’s t test)
Expression of miR-17, miR-20a and miR-25 significantly increased at embryonic day 15 (E15.5) relative to their expression earlier in pancreas development (Fig. 1c), consistent with their reported role in cell proliferation and tissue growth. To evaluate the expression levels of the miR-17-92 clusters in adult replicating beta cells, we obtained sorted beta cells from CcnB1-GFP transgenic mice, which express eGFP in replicating beta cells [25, 26]. mRNA levels of Ki67 (also known as Mki67) and the gene encoding DNA topoisomerase II α (Top2a) were upregulated in the sorted cells, confirming that this population is indeed in the cell division cycle (Fig. 1d). The expression of miR-17-92 and miR-106b-25 clusters was upregulated in proliferating cells, while the expression of other miRNAs (miR-375, miR-127) was unchanged (Fig. 1e). Therefore, miR-17-92/miR-106b-25 are induced in dividing beta cells.
miR-17-92/miR-106b-25 involvement in endocrine function
To study miR-17-92/miR-106b-25 family function in the mouse pancreas, we crossed mice carrying the miR-17-92 conditional allele with Pdx1-Cre transgenic mice and further mated the pedigree to mice carrying the miR-106b-25 whole-body KO allele [13] (Fig. 2a). The Pdx1-Cre; miR-17-92LoxP/LoxP;miR-106-25−/− cross resulted in significant downregulation of miR-17, miR-25 and miR-20a in islets, relative to control mice (harbouring miR-17-92LoxP/LoxP;miR-106-25+/− alleles; Fig. 2b).
Pancreas-specific loss of miR-17-92/miR-106b-25 impairs endocrine function. (a) Diagram of mouse genetics: miR-17-92LoxP/LoxP;miR-106b-25+/− (control) vs Pdx1-Cre;miR-17-92LoxP/LoxP;miR-106b-25−/− (miR-17-92/miR-106b-25-KO). (b) Relative expression levels of miR-17, miR-20a and miR-25 in 6-week-old KO and control mouse islets. Expression is normalised to the expression level in control tissue and to the expression of the non-coding RNA U6. n=3 mice. (c) GTT at multiple time points after glucose i.p. bolus (2 mg/g) in 6-month-old mice. miR-17-92/miR-106b-25-KO mice develop glucose intolerance, relative to control littermates. n=15 mice. (d) Area under the GTT curve shows an increase in the integrated sum of the blood glucose levels in the miR-17-92/miR-106b-25-KO vs control mice. (e) Representative micrograph of insulin immunohistochemistry in control and miR-17-92/miR-106b-25-KO mouse pancreases counter-stained with haematoxylin. Scale bar, 1000 μm. (f) Beta cell mass calculated as the fraction of insulin-positive area in consecutive sections spanning the entire pancreas (approximately 35 sections/pancreas) and multiplied by the pancreas mass in 4-month-old mice. n=5 mice. (g) ELISA measurement of insulin after ethanol extraction from whole pancreases of 4-month-old mice. n=5 mice. (h) Plasma insulin levels in peripheral blood before (0) or 15 min after i.p. injection of glucose (2 mg/kg) revealed reduced insulin secretion in 4- to 6-week-old KO mice relative to control mice. n=7 (control) or 10 (KO) mice. (i) Insulin secretion in isolated cultured islets of Langerhans, 60 min after low (2.5 mmol/l) or high (20 mmol/l) glucose stimulation, relative to total islet insulin. KO islets exhibit impaired response to glucose stimulus. n=5 mice. Data are presented as mean ± SEM. Except for (h) and (i), grey circles and bars, control mice; purple squares and bars, miR-17-92/miR-106b-25-KO mice. Significance was assessed with two-sided Student’s t test. *p<0.05, **p<0.01 and ***p<0.001. For (c) significance was calculated for the comparison between KO and control mice at different time points. miR-17-92cond, miR-17-92conditional; miR-106b het, miR-106-25 heterozygous; miR-106b KO, miR-106b-25 KO
We performed GTTs on four intermediate genotypes (ESM Fig. 1a), revealing an additive role for miR-17-92 and miR-106b-25 clusters in glucose homeostasis. Complete nullification resulted in the most severe impairment in glucose tolerance. We therefore investigated mutant mice lacking miR-17-92 and miR-106b-25 (Pdx1-Cre;miR-17-92LoxP/LoxP;miR-106-25−/−, referred to as miR-17-92/miR-106b-25-KO) vs littermate controls (miR-17-92LoxP/LoxP;miR-106-25+/−). Impaired glucose tolerance was evident at 3 months of age and progressively deteriorated at 6 and 12 months (Fig. 2c,d and ESM Fig. 1b), comparable with the results of Chen et al [39]. An ITT demonstrated normal response to insulin in mutant and control mice (ESM Fig. 1c), indicating that whole-body miR-106b-25-KO does not cause insulin resistance in peripheral tissues under these experimental conditions.
Morphometric analysis (Fig. 2e) revealed reduced beta cell mass in miR-17-92/miR-106b-25-KO vs control mouse pancreases (Fig. 2f). Moreover, there was a 50% decrease in total pancreatic insulin content in miR-17-92/miR-106b-25-KO vs control mice (Fig. 2g).
Immediately after i.p. injection of glucose, serum insulin levels were significantly diminished in miR-17-92/miR-106b-25-KO vs control mice (Fig. 2h). To distinguish between an intrinsic insulin secretion defect and a secondary effect due to reduced beta cell mass, we performed ex vivo GSIS tests on islets isolated from miR-17-92/miR-106b-25-KO mice or littermate control mice. Insulin secretion from isolated miR-17-92/miR-106b-25-KO mouse islets was diminished, even when normalised to insulin content (Fig. 2i). We therefore conclude that, in addition to controlling beta cell mass, miR-17-92/miR-106b-25 are required autonomously in beta cells for normal GSIS.
To characterise insulin secretion further, we isolated islets and performed GSIS in a perifusion apparatus. Perifused islets from miR-17-92/miR-106b-25-KO mice secreted insulin in a manner comparable with control islets in low glucose, but failed to display enhanced insulin secretion in response to high glucose, (Fig. 3a,b). The temporal secretion pattern involved both early (first) and delayed (second) phases, with a notable >50% reduction in insulin secretion in the miR-17-92/miR-106b-25-KO vs control mouse islets (Fig. 3a). However, forced-depolarisation of miR-17-92/miR-106-25-KO mouse beta cells with the non-nutrient secretagogue KCl produced a response comparable with that of control islets (Fig. 3a,b). These results demonstrate that miR-17-92/miR-106b-25 impact islet insulin secretion via a mechanism acting upstream of the plasma membrane depolarisation. Finally, we tested insulin secretion in the presence of cytochalasin B, a cell-permeable c-mycotoxin, which inhibits actin polymerisation and thus increases GSIS [40]. Cytochalasin B normalised insulin secretion in miR-17-92/miR-106b-25-KO mouse islets (Fig. 3c,d), suggesting that the capacity to synthesise insulin and assemble it into secretory granules is maintained in miR-17-92/miR-106b-25-KO mouse beta cells, while the regulated secretion pathway is impaired at a position upstream of potassium-dependent cell membrane depolarisation.
miR-17-92/miR-106b-25 are necessary for continuous GSIS. (a) GSIS of control vs miR-17-92/miR-106b-25-KO islets in a perifusion apparatus under conditions of low (2.8 mmol/l) or high (16.7 mmol/l) glucose concentration, or when depolarised with KCl (30 mmol/l). (b) Quantification of secreted insulin integral as AUC for the first and second phases of secretion (45–52 min and 52.3–95 min, respectively) and in response to KCl. (c) Continuous GSIS with cytochalasin B (CB, 10 μmol/l) under conditions as in (a). (d) Comparable insulin secretion by control and miR-17-92/miR-106b-25-KO islets in response to cytochalasin B. AUC of secreted insulin for the whole perifusion period (45–95 min). Islets from 3 mice, 4- to 6-week-old, per experimental condition. Statistical significance was calculated in (a) and (c) for the comparison between KO and control mice at different time points with two-way ANOVA followed by Sidak’s multiple comparison test. In (b) and (d), data are presented as mean ± SEM. Significance was assessed with two‐sided Student’s t test. *p<0.05, **p<0.01 and ***p<0.001; †p<0.001 in (a). Grey circles and bars, control mice; purple squares and bars, miR-17-92/miR-106b-25-KO mice
miR-17-92/miR-106b-25 KO do not affect beta cell apoptosis
Reduced beta cell mass could result from either a defect in proliferation or from beta cell apoptosis, consistent with the impact of miR-17-92/miR-106b-25 on proliferation and apoptosis in other tissues [22]. miR-17-92/miR-106b-25 family members are suppressors of the proapoptotic genes BIM (also known as BCL2L11) and PTEN [41], suggesting that an increase in apoptosis may occur, when miR-17-92/miR-106b-25 genes are deleted. Apoptosis was neither detected with activated caspase 3 nor with TUNEL staining of pancreas sections from mice aged 4 weeks or 12 months (ESM Fig. 2), in accordance with similar data from [39]. Therefore, it is likely that miR-17-92/miR-106b-25 family members regulate beta cell mass via proliferation rather than beta cell apoptosis.
miR-17-92/miR-106b-25 regulate beta cell proliferation
To test directly the contribution made by miR-17-92/miR-106b-25 to the proliferation of beta cells, we examined the expression levels of cell-cycle-related genes in mutant mice. Both nuclear markers of proliferation Ki-67 and DNA topoisomerase II α (encoded by Top2a), serve as sensitive proliferation markers in the endocrine pancreas [42, 43]. Ki67 and Top2a mRNA levels were downregulated in miR-17-92/miR-106b-25-KO vs control mouse islets (Fig. 4a) and the percentage of Ki-67-positive beta cells was also reduced (Fig. 4b). Therefore, miR-17-92/miR-106b-25 activity is important for beta cell proliferation.
Impaired beta cell proliferation in miR-17-92/miR-106b-25-KO mice. (a) Ki67 and Top2a mRNA expression levels in islets isolated from miR-17-92/miR-106b-25-KO mice or control littermates. n=7 (Ki67) or n=4 (Top2a). (b) Quantification of beta cells, which are positive for insulin and Ki67 immunostaining, in sections of miR-17-92/miR-106b-25-KO and control mouse pancreases (n=5 mice per group, >60 islets per mouse). (c) Diagram of the experimental design, depicting BrdU pulse and a chase of either 2 h or 36 h. (d) The percentage of BrdU-positive beta cells is comparable in miR-17-92/miR-106b-25-KO and control mouse islets 2 h post BrdU injection. (e) The percentage of BrdU-positive beta cells is reduced at 36 h post injection in miR-17-92/miR-106b-25-KO vs control mouse islets. n=3 mice per condition, >60 islets per mouse, >5000 beta cells counted per mouse. Data are shown as individual values and as mean ± SEM, *p<0.05 (two-sided Student’s t test)
Next, we hypothesised that miR-17-92 family members affect the ability of beta cells to enter or to successfully complete the cell division cycle. We quantified the percentage of beta cells engaged in DNA synthesis by BrdU labelling and determined the percentage of BrdU-positive beta cells out of total insulin-positive cells ([44], diagram in Fig. 4c). BrdU incorporation at 2 h post-injection represents the fraction of beta cells engaged in the S phase (DNA synthesis) at the time of the study (Fig. 4d). Unexpectedly, we observed a similar proportion of BrdU-positive beta cells in control and miR-17-92/miR-106b-25-KO mouse pancreases, suggesting that miR-17-92 family members are not essential for beta cell DNA synthesis.
To discover whether miR-17-92/miR-106b-25-KO mouse beta cells fail to accomplish the cell division cycle, we extended the study to 36 h after BrdU incorporation (i.e. longer than the full beta cell cycle [45]). After accomplishment of cytokinesis, the fraction of BrdU-positive wild-type beta cells doubled (from ~2% at 2 h to ~4% at 36 h, Fig. 4d,e), consistent with the expected doubling of the cell population. However, the fraction of BrdU-positive beta cells in miR-17-92/miR-106b-25-KO mouse islets remained unchanged (~2% at 2 h and 36 h), suggesting a severe defect in the cell division cycle. The defect was observed in islets of all sizes (ESM Fig. 3). Therefore, miR-17-92 is required for beta cells to effectively proceed through the cell cycle, at a point that is later than DNA synthesis, in contrast to miR-17-92 family activity described in other contexts. [13].
miR-17-92/miR-106b-25-KO mouse beta cells are delayed in the mitotic checkpoint
Because it appears that G1/S transition is not the main target of miR-17-92/miR-106b-25 in beta cells, we tested whether later checkpoints, G2/M and M, are regulated. We performed dual labelling of PHH3, a marker of M phase [45], along with BrdU. The transition through cell cycle checkpoints is disclosed by calculating the fraction of the total PHH3-positive beta cells that are positive for both BrdU and PHH3 (Fig. 5a). The BrdU+PHH3+ / PHH3+ ratio post-BrdU pulse revealed that the fraction of cells undergoing mitosis (M) and engaged in DNA synthesis (S) at the 3 h time window of the experimental chase was comparable in miR-17-92/miR-106b-25-KO and control beta cells. This suggests that miR-17-92/miR-106b-25 do not contribute to beta cell G2/M checkpoint dynamics (Fig. 5b).
miR-17-92/miR-106b-25-KO mouse beta cells are delayed in mitosis. (a) Diagram of cells progressing through the cell cycle. When BrdU is injected, cells (2) and (3) are at the DNA synthesis stage and incorporate BrdU, while most cells in the tissue (1) are unlabelled. Later, some cells exit S and retain BrdU (2), progress to mitosis and are co-labelled with PHH3 (3). A delay in transition through the G2/M checkpoint may be observed as relative depletion of PHH3+BrdU+ cells at 3 h, whereas a delay in M checkpoint will result in accumulation of PHH3+BrdU+ cells at 18 h. (b) The percentage of PHH3+BrdU+ cells (expressed as a percentage of the total PHH3+ cells) 3 h after BrdU injection was comparable in miR-17-92/miR-106b-25-KO and littermate control mouse beta cells. (c) The percentage of PHH3+BrdU+ cells 18 h after BrdU injection was doubled in miR-17-92/miR-106b-25-KO vs control mouse beta cells, suggesting a delay at the M checkpoint. Mice aged 4–6 weeks were used. Data are shown as individual values and as mean ± SEM. n=3. **p<0.01 (two-sided Student’s t test)
We next addressed the hypothesis that miR-17-92/miR-106b-25 regulate the M phase checkpoint by a longer (18 h) chase period after BrdU labelling, as in [45]. At this point, normal beta cells, which were initially at the S phase, have already accomplished cytokinesis and accordingly downregulated PHH3. Unexpectedly, we observed an increase in the percentage of double-stained miR-17-92/miR-106b-25-KO vs control mouse beta cells (Fig. 5c). We interpret this as evidence in support of miR-17-92/miR-106b-25 activity at the mitotic checkpoint. To test the hypothesis that miR-17-92/miR-106b-25-KO mouse beta cells fail the mitotic checkpoint and accumulate in a tetraploid state, we quantified X-inactive specific transcript, a long non-coding RNA that coats one X-chromosome in female cells. The smFISH study of Xist signal, used to detect duplicated X-chromosomes in tetraploid cells, did not reveal an increase in the abundance of tetraploid cells in miR-17-92/miR-106b-25-KO vs control mouse beta cells (ESM Fig. 4a). Therefore, we could not support the hypothesis that miR-17-92/miR-106b-25-KO cells undergo polyploidisation.
Transcriptome and proteome analysis of miR-17-92/miR-106b-25-KO cells
To discover relevant miR-17-92/miR-106b-25 targets, we profiled islet mRNA by next generation sequencing (NGS), differential mRNA expression and gene ontology analysis [46]. This depicted terms related to hormone (insulin) secretion and to cell cycle regulation among the over-represented biological pathways (Fig. 6a).
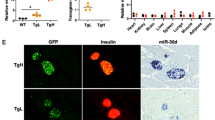
Integrated transcriptome and proteome analyses. (a) Biological pathway over-representation analysis of the genes with significantly altered expression in the miR-17-92/miR-106b-25-KO islets, depicting pathways involved in regulation of secretion, cell cycle and kinase activity. The red line indicates a p value of 0.05. (b) The expression of miR-17-92/miR-106b-25 family members in MEFs is reduced upon transduction with the CRE-adenovirus compared with control GFP-adenovirus. n=4. (c) Expression of cell cycle markers is downregulated in MEFs upon loss of miR-17-92/miR-106b-25. n=3. (d) The percentage of MEFs engaged in the cell cycle is reduced in MEFs infected with CRE- compared with control GFP-adenovirus. n=4. (e) Volcano plot of the proteins downstream of miR-17-92/miR-106b-25 KO. (f) MA-plot for differential analysis of the mass spectrometry samples. (g) PANTHER analysis of the proteins that were significantly different in the miR-17-92/miR 106b-25-KO MEFs compared with control MEFs reveals enrichment in biological pathways related to cytoskeleton organisation, regulation of mitosis and cell cycle phase transitions. The red line indicates a p value of 0.05. (h) 664 mRNA species were upregulated (Up) in miR-17-92/miR-106b-25-KO mouse islets, relative to control. 1258 proteins were upregulated in miR-17-92/miR-106b-25-KO MEFs (determined by MS). Of these, only four genes were upregulated with a corresponding effect at the protein level and are also predicted miR-17-92/miR-106b-25 targets from TargetScan. n=6 MEFs for each condition, corrected p value <0.05. Data are presented as individual values and as mean ± SEM. *p<0.05, **p<0.01 and ***p<0.001 (two-sided Student’s t test)
Bulk analysis in primary beta cells is limiting because only a small fraction of the beta cells are engaged with the cell division cycle. Therefore, we studied miR-17-92/miR-106b-25 activity in MEFs (miR-17-92LoxP/LoxP;miR-106-25−/− MEFs) derived from the same mouse allele and transduced with either GFP-adenovirus (control) or CRE-adenovirus. Comparing miRNA activity in endocrine pancreas and fibroblasts is biased towards detection of proteins that are expressed in both cell types and thus overlooks cell-type-specific expression. However, Ki67 and Top2a were downregulated in miRNA-deficient MEFs, reminiscent of miR-17-92/miR-106b-25-KO beta cells (Fig. 6b,c). Furthermore, the percentage of Ki-67-positive cells in MEFs depleted of miRNA was reduced relative to control MEFs (Fig. 6d).
MS analysis was performed on miR-17-92LoxP/LoxP;miR-106-25−/− MEF lysate, without or with CRE-Adenovirus. miR-17-92LoxP/LoxP;miR-106-25+/+ MEFs with GFP-adenovirus served as a control. Comparable results were gained when miR-17-92LoxP/LoxP;miR-106-25−/− MEFs with GFP-adenovirus were used as controls. MS in six experimental repeats depicted 16,005 unique peptides, corresponding to 2715 different proteins. The expression level of 64% of the proteins was significantly changed by knocking out miR-17-92/miR-106b-25. The majority of significantly changed proteins were upregulated (84.6%, Fig. 6e,f). Intriguingly, mitosis and cell cycle regulation were among the significantly enriched GO terms along with cytoskeleton organisation and actin filament polymerisation regulation (Fig. 6g). Out of the 1258 upregulated proteins, 146 were predicted targets of at least one miRNA from the miR-17-92/miR-106b-25 family (Fig. 6h). Fifty-six gene products were significantly upregulated at both the mRNA (in islets) and protein (in fibroblasts) levels. Four out of these 56 were predicted direct targets of at least one member of the miR-17-92/miR-106b-25 family, making a short list of highly relevant targets across tissues. These targets include Mark2 encoding microtubule affinity regulating kinase 2 (MARK2), Jpt1/Hn1 encoding Jupiter microtubule associated homolog 1, Sqstm1 encoding sequestosome 1 and Prkar1a encoding protein kinase cAMP-dependent type I regulatory subunit α (PRKAR1α).
Both MARK2 and PRKAR1α are part of the protein kinase A (PKA) pathway, the cellular sensor of cAMP, which regulates cell division cycle and insulin secretion. [47,48,49]. Prkar1a levels were validated by qRT-PCR (Fig. 7a). Prkar1a and Mark2 upregulation suggests a potential role for the PKA pathway downstream of miR-17-92. We therefore studied the hypothesis that PKA activity is reduced in the islets isolated from miR-17-92/miR-106b-25-KO mice. PKA activity was downregulated in miR-17-92/miR-106b-25-KO vs control littermate mouse islets. Furthermore, islets harbouring the miR-17-92 overexpression transgene Pdx-Cre;ROSA-miR-17-92conditional displayed elevated PKA activity (Fig. 7b and ESM Fig. 4b). Taken together, PKA activity is sensitive bidirectionally to miR-17-92/miR-106b-25 levels in beta cells. Control of PKA by miR-17-92/miR-106b-25 is a new convergence point for seemingly disparate processes of proliferation and insulin secretion.
PKA activity and suggested model downstream of miR-17-92/miR-106b-25. (a) Prkar1a expression is elevated in isolated miR-17-92/miR-106b-25-KO mouse islets relative to control littermates. n=4. (b) PKA activity is reduced in miR-17-92/miR-106b-25-KO mouse islets, while it is elevated in islets harbouring the miR-17-92 overexpression transgene (Pdx-Cre;ROSA-miR-17-92conditional [OE] and ROSA-miR-17-92conditional [Control OE]). Data are shown as individual values and mean ± SEM. n=4. *p<0.05 and ***p<0.001 (two-sided Student’s t test). (c) Suggested model for the regulation of the mitotic checkpoint and insulin secretion downstream of miR-17-92/miR-106b-25. PRKAR1α, the most abundant regulatory subunit of the PKA holoenzyme complex, keeps the complex in an inactive form. Upon release, the PKA catalytic subunit can phosphorylate its downstream targets, thus contributing to microtubule stabilisation while preventing accumulation of cells in the G2 and M stages of the cell cycle. Moreover, different phosphorylated downstream targets promote insulin exocytosis. (d) miR-17-92/miR-106b-25 expression is downregulated when beta cells mature, correlating with a reduction in beta cell proliferation. Insulin exocytosis is inhibited by miR-17-92 in immature beta cells [20], whereas we demonstrated that miR-17-92 and miR-106b-25 induce GSIS in mature beta cells. Changes in miRNA levels, or changes in the functions of specific mRNA targets that are differentially expressed in immature vs mature beta cells, might contribute to explaining this discrepancy. EPAC2, exchange protein directly activated by cAMP2; SNAP25, synaptosome associated protein 25; Snapin, SNAP associated protein
Discussion
Using mouse genetics we discovered that miR-17-92/miR-106b-25 alleles regulate islet function via control of beta cell mass and insulin secretion. The miR-17-92/miR-106b-25 family is important for normal endocrine function and, accordingly, loss of the miRNAs results in endocrine failure.
miR-17-92/miR-106b-25-KO mice exhibited normal insulin tolerance and miR-106b-25 deficiency in insulin-responsive tissues (muscle, liver and adipose) did not modify target organ insulin sensitivity, in vivo. Therefore, endocrine pancreas failure manifesting as a reduction in GSIS is a plausible cause of glucose intolerance.
Our study suggests a more pronounced role for miR-17-92 in GSIS than was previously reported [39], in part, since nullification of both miR-17-92 and miR-106b-25 clusters in our model reduces the overall miRNA levels from these clusters, relative to the reduction achieved by KO of just the miR-17-92 cluster. Additional GSIS study in a perifusion apparatus revealed that miR-17-92/miR-106b-25 are involved in insulin secretion at a stage preceding membrane depolarisation. Accordingly, non-nutrient secretagogue KCl normalised insulin exocytosis, suggesting that voltage-dependent calcium channels and successive events, such as granule docking and fusion, are insensitive to miR-17-92/miR-106b-25. Likewise, a study with cytochalasin B revealed that insulin synthesis, granule assembly, docking and fusion are unaffected by miR-17-92/miR-106b-25. Taken together, these analyses delineate miR-17-92/miR-106b-25 activity at steps earlier than membrane depolarisation.
Several studies proposed that miR-17-92/miR-106b-25 upregulation is important for S phase entry in transformed cell lines and tissues [16, 50], whereas our work uncovers a new role for miR-17-92/miR-106b-25 in regulation of the M checkpoint. The involvement in the M checkpoint might have been overlooked until now or alternatively could reflect different miR-17-92/miR-106b-25 functions in genetically stable tissues. Interestingly, miR-17-92 expression is highest during G2/M and lowest in S phase even in transformed cells [16].
The final consequences of miR-17-92/miR-106b-25 deficiency in adult replicating beta cells are unclear, since we observed neither increased apoptosis nor accumulation of polyploid beta cells in young or old miR-17-92/miR-106b-25-KO mouse islets. Thus, the most likely explanation is that dividing adult miR-17-92/miR-106b-25-KO mouse beta cells undergo non-apoptotic cell death and rapid tissue clearance at a rate higher than our assay sensitivity.
c-Myc (also known as Myc) overexpression in beta cells induces cell cycle and reduces insulin expression and apoptosis [22]. miR-17-92/miR-106b-25 might act as c-Myc effectors in mediating at least some of its functions, as occurs in some cancers. In addition, c-Myc resides in a positive feedback loop with PKA [51, 52]. Therefore, miR-17-92/miR-106b-25 family, c-Myc and PKA activities may be interwoven.
High cAMP levels induce insulin secretion via PKA- and Epac2 (also known as Rapgef4)-dependent recruitment of insulin granules and/or granule fusion to the plasma membrane [53, 54]. PRKAR1α inhibits insulin secretion, whereas cAMP antagonises PRKAR1α and releases PKA from PRKAR1α subunit. This way, cAMP facilitates PKA-dependent induction of Snapin–Snap25–Epac2 pathway activity, resulting in increased insulin exocytosis [55, 56]. In silencing Prkar1a, miR-17-92/miR-106b-25 induce PKA activity and insulin exocytosis. Furthermore, PKA regulates microtubule stability and potentially the M checkpoint by phosphorylating Mark2 on S409 [57,58,59].
Therefore, PKA is a convergence point that contributes both to insulin exocytosis and to mitotic checkpoint, connecting two seemingly disparate properties, namely beta cell division and insulin secretion, downstream of miR-17-92/miR-106b-25 family (Fig. 7c). However, our current study does not provide direct experimental evidence that connects microtubule stability to PKA activity in miR-17-92/miR-106b-25-KO mice.
More broadly, miR-17-92/miR-106b-25 activity may be a new regulatory element, contributing to activity of incretin-stimulated pathways via PKA and to some of the therapeutic actions of glucagon-like peptide-1 (GLP-1) on insulin exocytosis and beta cell proliferation.
We suggest that in adult mice miR-17-92/miR-106b-25 expression is upregulated significantly but transiently in dividing beta cells, contributing to beta cell proliferation via a mechanism similar to that reported in early postnatal maturation [20]. Mature beta cells express only low miR-17-92/miR-106b-25 levels, which primarily affect insulin secretion (Fig. 7d).
Overall, our study deciphers the involvement of miR-17-92/miR-106b-25 family in adult beta cell replication and insulin secretion, suggesting an important role for proto-oncogene miRNAs in regulating glucose homeostasis in the normal, untransformed endocrine pancreas. In mice, miR-17-92/miR-106b-25 appear to regulate many facets of the adult beta cell life, connecting mitosis and insulin secretion by a single post-transcriptional pathway, encouraging similar studies in human beta cells.
Data availability
Sequencing data that support the findings of this study have been deposited in GEO with the accession codes GSE126516 [34].
Abbreviations
- BrdU:
-
Bromodeoxyuridine
- FDR:
-
False discovery rate
- FRET:
-
Fluorescence resonance energy transfer
- GO:
-
Gene ontology
- GSIS:
-
Glucose-stimulated insulin secretion
- KO:
-
Knockout
- MARK2:
-
Microtubule affinity regulating kinase 2
- MEF:
-
Mouse embryonic fibroblast
- miRNA:
-
MicroRNA
- PHH3:
-
Phosphorylated histone H3
- PKA:
-
Protein kinase A
- PRKAR1α:
-
Protein kinase cAMP-dependent type I regulatory subunit α
- qRT-PCR:
-
Quantitative real-time RT-PCR
- smFISH:
-
Single molecule fluorescence in situ hybridisation
References
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297. https://doi.org/10.1016/S0092-8674(04)00045-5
Lynn FC (2009) Meta-regulation: microRNA regulation of glucose and lipid metabolism. Trends Endocrinol Metab 20(9):452–459. https://doi.org/10.1016/j.tem.2009.05.007
Joglekar MV, Parekh VS, Hardikar AA (2011) Islet-specific microRNAs in pancreas development, regeneration and diabetes. Indian J Exp Biol 49(6):401–408
Walker MD (2008) Role of MicroRNA in pancreatic beta-cells: where more is less. Diabetes 57(10):2567–2568. https://doi.org/10.2337/db08-0934
Melkman-Zehavi T, Oren R, Kredo-Russo S et al (2011) miRNAs control insulin content in pancreatic beta-cells via downregulation of transcriptional repressors. EMBO J 30(5):835–845. https://doi.org/10.1038/emboj.2010.361
Mandelbaum AD, Melkman-Zehavi T, Oren R et al (2012) Dysregulation of Dicer1 in beta cells impairs islet architecture and glucose metabolism. Exp Diabetes Res 2012:470302
Ackermann AM, Gannon M (2007) Molecular regulation of pancreatic beta-cell mass development, maintenance, and expansion. J Mol Endocrinol 38(1–2):193–206. https://doi.org/10.1677/JME-06-0053
Bernard-Kargar C, Ktorza A (2001) Endocrine pancreas plasticity under physiological and pathological conditions. Diabetes 50(Suppl 1):S30–S35. https://doi.org/10.2337/diabetes.50.2007.S30
Meier JJ, Butler AE, Saisho Y et al (2008) Beta-cell replication is the primary mechanism subserving the postnatal expansion of beta-cell mass in humans. Diabetes 57(6):1584–1594. https://doi.org/10.2337/db07-1369
Dor Y, Brown J, Martinez OI, Melton DA (2004) Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature 429(6987):41–46. https://doi.org/10.1038/nature02520
Nurse P (1990) Universal control mechanism regulating onset of M-phase. Nature 344(6266):503–508. https://doi.org/10.1038/344503a0
Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL (2007) Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol 310(2):442–453. https://doi.org/10.1016/j.ydbio.2007.08.007
Ventura A, Young AG, Winslow MM et al (2008) Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132(5):875–886. https://doi.org/10.1016/j.cell.2008.02.019
Pelengaris S, Khan M, Evan GI (2002) Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell 109(3):321–334. https://doi.org/10.1016/S0092-8674(02)00738-9
Mogilyansky E, Rigoutsos I (2013) The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ 20(12):1603–1614. https://doi.org/10.1038/cdd.2013.125
Cloonan N, Brown MK, Steptoe AL et al (2008) The miR-17-5p microRNA is a key regulator of the G1/S phase cell cycle transition. Genome Biol 9(8):R127. https://doi.org/10.1186/gb-2008-9-8-r127
O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT (2005) c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435(7043):839–843. https://doi.org/10.1038/nature03677
Sylvestre Y, De Guire V, Querido E et al (2007) An E2F/miR-20a autoregulatory feedback loop. J Biol Chem 282(4):2135–2143. https://doi.org/10.1074/jbc.M608939200
Petrocca F, Visone R, Onelli MR et al (2008) E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 13(3):272–286. https://doi.org/10.1016/j.ccr.2008.02.013
Jacovetti C, Matkovich SJ, Rodriguez-Trejo A, Guay C, Regazzi R (2015) Postnatal beta-cell maturation is associated with islet-specific microRNA changes induced by nutrient shifts at weaning. Nat Commun 6(1):8084. https://doi.org/10.1038/ncomms9084
Jacovetti C, Rodriguez-Trejo A, Guay C et al (2017) MicroRNAs modulate core-clock gene expression in pancreatic islets during early postnatal life in rats. Diabetologia 60(10):2011–2020. https://doi.org/10.1007/s00125-017-4348-6
Laybutt DR, Weir GC, Kaneto H et al (2002) Overexpression of c-Myc in beta-cells of transgenic mice causes proliferation and apoptosis, downregulation of insulin gene expression, and diabetes. Diabetes 51(6):1793–1804. https://doi.org/10.2337/diabetes.51.6.1793
Hingorani SR, Petricoin Iii EF, Maitra A et al (2003) Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 4(6):437–450. https://doi.org/10.1016/S1535-6108(03)00309-X
Xiao C, Srinivasan L, Calado DP et al (2008) Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol 9(4):405–414. https://doi.org/10.1038/ni1575
Klochendler A, Caspi I, Corem N et al (2016) The Genetic Program of Pancreatic beta-Cell Replication In Vivo. Diabetes 65(7):2081–2093. https://doi.org/10.2337/db16-0003
Klochendler A, Weinberg-Corem N, Moran M et al (2012) A transgenic mouse marking live replicating cells reveals in vivo transcriptional program of proliferation. Dev Cell 23(4):681–690. https://doi.org/10.1016/j.devcel.2012.08.009
Noordeen NA, Khera TK, Sun G et al (2010) Carbohydrate-responsive element-binding protein (ChREBP) is a negative regulator of ARNT/HIF-1beta gene expression in pancreatic islet beta-cells. Diabetes 59(1):153–160. https://doi.org/10.2337/db08-0868
Durkin ME, Qian X, Popescu NC, Lowy DR (2013) Isolation of Mouse Embryo Fibroblasts. Bio Protoc 3(18):e908
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14(4):R36. https://doi.org/10.1186/gb-2013-14-4-r36
Anders S, Pyl PT, Huber W (2015) HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31(2):166–169. https://doi.org/10.1093/bioinformatics/btu638
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):550. https://doi.org/10.1186/s13059-014-0550-8
Huang DA, Sherman WBT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4(1):44–57. https://doi.org/10.1038/nprot.2008.211
Huang DA, Sherman WBT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37(1):1–13. https://doi.org/10.1093/nar/gkn923
Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30(1):207–210. https://doi.org/10.1093/nar/30.1.207
Perez-Riverol Y, Csordas A, Bai J et al (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47(D1):D442–D450. https://doi.org/10.1093/nar/gky1106
Itzkovitz S, van Oudenaarden A (2011) Validating transcripts with probes and imaging technology. Nat Methods 8(4 Suppl):S12–S19. https://doi.org/10.1038/nmeth.1573
Bahar Halpern K, Itzkovitz S (2016) Single molecule approaches for quantifying transcription and degradation rates in intact mammalian tissues. Methods 98:134–142. https://doi.org/10.1016/j.ymeth.2015.11.015
Sommer C, Straehle C, Kothe U, Hamprecht FA (2011) Ilastik: Interactive Learning and Segmentation Toolkit. 2011 8th IEEE International Symposium on Biomedical Imaging: From Nano to Macro: 230–233
Chen Y, Tian L, Wan S et al (2016) MicroRNA-17-92 cluster regulates pancreatic beta-cell proliferation and adaptation. Mol Cell Endocrinol 437:213–223. https://doi.org/10.1016/j.mce.2016.08.037
Kong X, Yan D, Sun J et al (2014) Glucagon-like peptide 1 stimulates insulin secretion via inhibiting RhoA/ROCK signaling and disassembling glucotoxicity-induced stress fibers. Endocrinology 155(12):4676–4685. https://doi.org/10.1210/en.2014-1314
Wang Y, Lee CG (2009) MicroRNA and cancer--focus on apoptosis. J Cell Mol Med 13(1):12–23. https://doi.org/10.1111/j.1582-4934.2008.00510.x
Milde-Langosch K, Karn T, Muller V et al (2013) Validity of the proliferation markers Ki67, TOP2A, and RacGAP1 in molecular subgroups of breast cancer. Breast Cancer Res Treat 137(1):57–67. https://doi.org/10.1007/s10549-012-2296-x
Stolovich-Rain M, Hija A, Grimsby J, Glaser B, Dor Y Pancreatic beta cells in very old mice retain capacity for compensatory proliferation. J Biol Chem 287(33):27407–27414. https://doi.org/10.1074/jbc.M112.350736
Oh YS, Shin S, Lee Y-J, Kim EH, Jun H-S (2011) Betacellulin-induced beta cell proliferation and regeneration is mediated by activation of ErbB-1 and ErbB-2 receptors. PLoS One 6(8):e23894. https://doi.org/10.1371/journal.pone.0023894
Hija A, Salpeter S, Klochendler A et al (2014) G0-G1 transition and the restriction point in pancreatic beta-cells in vivo. Diabetes 63(2):578–584. https://doi.org/10.2337/db12-1035
Mi H, Muruganujan A, Casagrande JT, Thomas PD (2013) Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 8(8):1551–1566. https://doi.org/10.1038/nprot.2013.092
Du WW, Yang W, Fang L et al (2014) miR-17 extends mouse lifespan by inhibiting senescence signaling mediated by MKP7. Cell Death Dis 5(7):e1355. https://doi.org/10.1038/cddis.2014.305
Prentki M, Matschinsky FM (1987) Ca2+, cAMP, and phospholipid-derived messengers in coupling mechanisms of insulin secretion. Physiol Rev 67(4):1185–1248. https://doi.org/10.1152/physrev.1987.67.4.1185
Yan L, Vatner DE, O’Connor JP et al (2007) Type 5 adenylyl cyclase disruption increases longevity and protects against stress. Cell 130(2):247–258. https://doi.org/10.1016/j.cell.2007.05.038
Hayashita Y, Osada H, Tatematsu Y et al (2005) A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res 65(21):9628–9632. https://doi.org/10.1158/0008-5472.CAN-05-2352
Padmanabhan A, Li X, Bieberich CJ (2013) Protein kinase A regulates MYC protein through transcriptional and post-translational mechanisms in a catalytic subunit isoform-specific manner. J Biol Chem 288(20):14158–14169. https://doi.org/10.1074/jbc.M112.432377
Wu KJ, Mattioli M, Morse HC 3rd, Dalla-Favera R (2002) c-MYC activates protein kinase A (PKA) by direct transcriptional activation of the PKA catalytic subunit beta (PKA-Cbeta) gene. Oncogene 21(51):7872–7882. https://doi.org/10.1038/sj.onc.1205986
Seino S, Shibasaki T (2005) PKA-dependent and PKA-independent pathways for cAMP-regulated exocytosis. Physiol Rev 85(4):1303–1342. https://doi.org/10.1152/physrev.00001.2005
Drucker DJ, Nauck MA (2006) The incretin system: glucagon-like peptide-1 receptor agonists and dipeptidyl peptidase-4 inhibitors in type 2 diabetes. Lancet 368(9548):1696–1705. https://doi.org/10.1016/S0140-6736(06)69705-5
Hussain MA, Stratakis C, Kirschner L (2012) Prkar1a in the regulation of insulin secretion. Horm Metab Res 44(10):759–765. https://doi.org/10.1055/s-0032-1321866
Song WJ, Seshadri M, Ashraf U et al (2011) Snapin mediates incretin action and augments glucose-dependent insulin secretion. Cell Metab 13(3):308–319. https://doi.org/10.1016/j.cmet.2011.02.002
Drewes G, Ebneth A, Preuss U, Mandelkow EM, Mandelkow E (1997) MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell 89(2):297–308. https://doi.org/10.1016/S0092-8674(00)80208-1
Deng SS, Wu LY, Wang YC et al (2015) Protein kinase A rescues microtubule affinity-regulating kinase 2-induced microtubule instability and neurite disruption by phosphorylating serine 409. J Biol Chem 290(5):3149–3160. https://doi.org/10.1074/jbc.M114.629873
Hubaux R, Thu KL, Vucic EA et al (2015) Microtubule affinity-regulating kinase 2 is associated with DNA damage response and cisplatin resistance in non-small cell lung cancer. Int J Cancer 137(9):2072–2082. https://doi.org/10.1002/ijc.29577
Acknowledgements
The authors would like to thank Y. Melamed and O. Higfa for veterinary services and husbandry (Weizmann Institute of Science). The authors thank A. Savidor and Y. Levin at the de Botton Institute for Protein Profiling, Weizmann Institute of Science, and O. Ben-Ami of the Crown Genomics Institute of the Nancy and Stephen Grand Israel National Center for Personalized Medicine, Weizmann Institute of Science, for MS and next generation sequencing, respectively. We thank O. Elhanani, M. Walker, S. Itzkovitz, H. Kaspi, R. Pasvolsky and E. Geron (Weizmann Institute of Science) for insightful comments on the manuscript.
Funding
The work is funded by an ERC consolidator program (617351), Juvenile Diabetes Research Foundation (99-2007-71 and 47-2012-742), European Diabetes Research Programmes (EFSD)/D-Cure young Investigator award and EFSD-Lilly, Yeda-Sela, Yeda-CEO, Y. Leon Benoziyo Institute for Molecular Medicine, Kekst Family Institute for Medical Genetics, David and Fela Shapell Family Center for Genetic Disorders Research, Crown Human Genome Center, Nathan, Shirley, Philip and Charlene Vener New Scientist Fund, Julius and Ray Charlestein Foundation, Fraida Foundation, Wolfson Family Charitable Trust, Adelis Foundation, Merck (UK), Maria Halphen and Estates of Fannie Sherr, Lola Asseof, Lilly Fulop. Hornstein laboratory is supported by Dr. Sydney Brenner. EH is Head of Nella and Leon Benoziyo Center for Neurological Diseases and incumbent of Ira & Gail Mondry Professorial chair. The study sponsor was not involved in the design of the study, the collection, analysis and interpretation of data, writing the report or the decision to submit the report for publication.
Author information
Authors and Affiliations
Contributions
ADM, SK and EH made substantial contributions to the conception or design of the work, the acquisition, analysis and interpretation of data and drafting the work for important intellectual content. YD provided substantial contribution to the conception or design of the work and drafting the work for important intellectual content and agrees to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. EY, AK, AS, NM and DA contributed to acquisition, analysis or interpretation of data and drafting the work for important intellectual content. All authors gave final approval of the version to be published. EH is the guarantor of this work.
Corresponding author
Ethics declarations
The authors declare that there is no duality of interest associated with this manuscript.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM Figs
(PDF 572 kb)
Rights and permissions
About this article
Cite this article
Mandelbaum, A.D., Kredo-Russo, S., Aronowitz, D. et al. miR-17-92 and miR-106b-25 clusters regulate beta cell mitotic checkpoint and insulin secretion in mice. Diabetologia 62, 1653–1666 (2019). https://doi.org/10.1007/s00125-019-4916-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-019-4916-z