Abstract
Aims/hypothesis
We aimed to determine the association of maternal metabolites with newborn adiposity and hyperinsulinaemia in a multi-ethnic cohort of mother–newborn dyads.
Methods
Targeted and non-targeted metabolomics assays were performed on fasting and 1 h serum samples from a total of 1600 mothers in four ancestry groups (Northern European, Afro-Caribbean, Mexican American and Thai) who participated in the Hyperglycemia and Adverse Pregnancy Outcome (HAPO) study, underwent an OGTT at ~28 weeks gestation and whose newborns had anthropometric measurements at birth.
Results
In this observational study, meta-analyses demonstrated significant associations of maternal fasting and 1 h metabolites with birthweight, cord C-peptide and/or sum of skinfolds across ancestry groups. In particular, maternal fasting triacylglycerols were associated with newborn sum of skinfolds. At 1 h, several amino acids, fatty acids and lipid metabolites were associated with one or more newborn outcomes. Network analyses revealed clusters of fasting acylcarnitines, amino acids, lipids and fatty acid metabolites associated with cord C-peptide and sum of skinfolds, with the addition of branched-chain and aromatic amino acids at 1 h.
Conclusions/interpretation
The maternal metabolome during pregnancy is associated with newborn outcomes. Maternal levels of amino acids, acylcarnitines, lipids and fatty acids and their metabolites during pregnancy relate to fetal growth, adiposity and cord C-peptide, independent of maternal BMI and blood glucose levels.
Similar content being viewed by others

Introduction
The developmental origins hypothesis posits that the intrauterine environment has an impact on fetal development and childhood health [1], since nutritional, metabolic and hormonal cues in utero can influence offspring susceptibility to disease. Specifically, newborn adiposity is a perinatal marker of adverse metabolic exposures in utero and has been linked to childhood adiposity, ultimately affecting future risk of metabolic diseases such as type 2 diabetes mellitus [2,3,4]. Maternal obesity and hyperglycaemia are well-known independent risk factors for newborn fat deposition, although excess adiposity at birth can be present even in their absence [5]. Understanding contributors to and biomarkers of newborn adiposity might help identify at-risk children early in life.
Metabolomics characterises metabolites that represent an individual’s biological status in a normal or pathological state. The metabolome is affected by genetic, dietary and environmental contributors; thus, metabolomics allows for a comprehensive understanding of pathophysiological states and provides mechanistic insight into disease development. During pregnancy, a mother’s metabolite profile represents her physiological state at a specific time point. Pre-pregnancy and intrapartum factors, such as BMI, gestational weight gain, body composition and maternal glucose, likely affect both a mother’s metabolite levels and newborn body composition, particularly newborn adiposity. A unique metabolic signature integrating several contributory pre-pregnancy and intrapartum factors might emerge as a proxy for the metabolic environment surrounding the fetus, which, in turn, might affect fetal organogenesis, fat deposition and insulin sensitivity.
Metabolomics has the potential to identify predictive markers of later adiposity and insulin resistance. In adults, branched-chain amino acids (BCAAs), aromatic amino acids (AAAs), short-chain acylcarnitines (C2, C6, C8; see ESM Table 1 for acylcarnitine nomenclature) and the carnitine ester of 3-hydroxybutyrate (acylcarnitine C4-OH) have all been associated with insulin resistance, higher HbA1c and/or postprandial glucose levels [5,6,7,8]. However, the impact of the maternal metabolome on the developing fetus, as evidenced by newborn outcomes, is largely unknown and understudied [9, 10].
Identification of a maternal metabolomic signature associated with adverse newborn outcomes might emerge as a way to determine risk early in life and inform targeted prevention efforts during pregnancy and early infancy. In this study of 1600 well-characterised, multi-ethnic mother–newborn pairs, we aimed to investigate associations of the maternal metabolome during pregnancy with newborn adiposity at birth to begin clarifying relationships between maternal metabolism and fetal development.
Methods
Data and sample collection
Maternal blood samples were obtained during a 75 g OGTT between 24 and 32 weeks gestation as part of the Hyperglycemia and Adverse Pregnancy Outcome (HAPO) Study [11]. Fasting, 1 h and 2 h glucose levels were measured as described [11]. Maternal samples were stored at −80°C prior to metabolomics assays. Metabolic profiling was performed on maternal fasting and 1 h serum samples from 400 mothers in each of four ancestry groups: Afro-Caribbean, Mexican American, Northern European and Thai.
Trained personnel obtained maternal anthropometrics using a standardised protocol at the time of the OGTT. Gestational age was calculated as previously described [11]. Demographic data were ascertained via questionnaire. Newborn anthropometric measurements, including birthweight and sum of skinfolds (SSF), were measured within 72 h of birth using calibrated equipment and standardised methods across field centres. The protocol was approved by the Institutional Review Board at all participating sites and each mother provided written informed consent for herself and her child.
Conventional metabolites and targeted metabolomics assays
Sixty-two conventional and targeted metabolites were assayed as previously described [12]. Briefly, conventional metabolite levels (lactate, triacylglycerols, 3-hydroxybutyrate, glycerol, NEFA) were measured on a Unicel DxC 600 clinical analyser (Beckman Coulter, Brea, CA, USA). Targeted metabolomics assays for acylcarnitines and amino acids were performed by tandem MS with addition of known quantities of stable isotope-labelled internal standards on an Acquity TQD Triple Quadrupole system (Waters Corporation, Milford, MA, USA).
Non-targeted analyses
Non-targeted GC–MS assays were performed to analyse the full range of metabolites in serum, as described [12]. Methanol, the extraction solvent, was spiked with a retention-time-locking (RTL) internal standard of perdeuterated myristic acid. Extracts were prepared for GC–MS by methoximation and trimethylsilylation [13, 14]. Sera from Northern European and Thai ancestries were run on 6890 N GC-5975B Inert MS (Agilent Technologies, Santa Clara, CA, USA). Afro-Caribbean and Mexican American samples were run on 7890B GC-5977B Inert MS (Agilent Technologies). Peaks were deconvoluted with AMDIS freeware [15] and parsed against the Fiehn RTL spectral library [14] with additions from our laboratory. Manual curation included identifying co-eluting groups of isomeric metabolites and selecting reliable peaks [13]. Detected peak areas were log2-transformed for analysis. In total, 73 GC–MS metabolites not assayed using targeted approaches were used for data analysis.
Fasting and 1 h maternal serum sample pairs were batched for GC–MS. Batches of equal size were run over 50 days and balanced by field centre, maternal glucose and BMI and newborn outcomes. Quality control (QC) pools were constructed using equal volumes from all maternal samples and prepared for analysis as described above. QCs were injected as first, middle and last samples of each GC–MS batch. To control technical variability attributable to batch and run order, GC–MS data were normalised using QC data and metabomxtr R package, version 1.16.0 [12, 16].
Statistical analysis
Per-metabolite analysis
Acylcarnitine and 3-hydroxybutyrate levels were log-transformed to improve normality. Outlying metabolite values, defined as values >5 SD from the mean, and data from individuals with more than ten outlying metabolites for a sample type (fasting, 1 h), were excluded prior to statistical analysis.
Associations between phenotypes and metabolites with <10% missing data within sample type were identified using linear regression within ancestry groups. Analyses treating maternal metabolites as predictors and newborn phenotypes as outcomes were adjusted for field centre, mean arterial pressure, maternal age, neonatal sex, sample storage time, maternal height and gestational age at delivery (model 1). Two additional models adjusted for the covariates above plus either maternal BMI at OGTT (model 2) or fasting or 1 h maternal glucose for the corresponding fasting or 1 h metabolite levels (model 3).
Meta-analysis
β values from per-metabolite analyses were combined across ancestry groups using random effects meta-analysis with inverse variance weights and restricted maximum likelihood estimation [17] using the ‘metafor’ R package, version 2.0.0 (http://www.metafor-project.org). Heterogeneity of effects among ancestry groups was assessed descriptively with I2 statistics and formally tested via Cochran Q tests. False discovery rate (FDR) correction was applied to meta-analysis and Cochran Q test p values, and FDR-adjusted p values <0.05 were considered statistically significant. All statistical analyses were conducted using R (version 3.4.3, http://www.r-project.org).
Network analyses
Graphical lasso techniques were used to identify metabolic networks associated with phenotype [18]. Metabolites are represented by nodes, and edges depict dependence of metabolite pairs conditional on all other metabolites. First, we identified the subset of metabolites associated with phenotype at nominal p < 0.05. Next, we identified all other metabolites demonstrating partial correlation with magnitude >0.25 with at least one significant metabolite after adjusting for model variables, as described previously. We calculated residuals from linear models for these metabolites including model covariates, and used these residuals as inputs into graphical lasso. Graphical lasso uses a coordinate descent algorithm and a penalty term to provide a sparse estimate of the inverse covariance matrix for a given set of input features. Zero covariance indicates independence for pairs of features conditional on all others, while non-zero covariance indicates dependence, conditional on all other features. In network parlance, if nodes represent input features (i.e. metabolites), non-zero covariance estimates correspond to edges between pairs of nodes that represent their association with each other, independent of all other graph features. The penalty term trims likely false-positive edges by gauging the strength of estimated edges.
Graphical lasso was applied within each ancestry group and in meta-analysis, using meta-analytical estimates of partial correlations based on a Fisher’s z-transformation. To further elucidate network structure, spinglass clusters [19] were estimated on graphical lasso networks to identify communities of nodes that are more tightly connected to each other than the other nodes in the network.
Results
Participant characteristics are shown in Table 1. All infants were born full-term and newborn sex was roughly equally distributed. Thai newborns had the lowest birthweight, while Thai and Afro-Caribbean newborns had the lowest SSF. Maternal glucose levels and BMI spanned the range observed in the HAPO study. Mexican American and Thai mothers had the highest fasting and 1 h glucose levels, respectively. Thai mothers had the lowest BMI and shortest stature.
Metabolites associated across ancestry groups
Figure 1 depicts 95% CIs for maternal fasting and 1 h metabolites that demonstrate significant associations with newborn birthweight, SSF and/or cord C-peptide in meta-analyses after FDR-adjustment in at least one model. The β estimates and FDR-adjusted p values are listed in ESM Table 2. The magnitude of the association between some metabolite levels and newborn outcomes was minimally affected by adjustment for maternal BMI and/or glucose, while for other metabolites, the magnitude and significance was greatly reduced.
Maternal metabolites associated with newborn outcomes across ancestries. Forest plot depicting maternal metabolites after fasting and at 1 h in the OGTT that are significantly associated with newborn birthweight (a), cord blood C-peptide (b) and/or newborn SSF (c). The x-axis represents the standardised β of each individual metabolite. Analyses were adjusted for field centre, mean arterial pressure, maternal age, neonatal sex, sample storage time, maternal height and gestational age at delivery (model 1). Model 2 included model 1 covariates and maternal BMI; model 3 included model 1 covariates and fasting or 1 h maternal glucose for the corresponding fasting or 1 h metabolite levels. Squares, model 1; circles, model 2; diamonds, model 3. AC, acylcarnitine
Fasting levels of few metabolites were associated with newborn outcomes. Maternal fasting triacylglycerols were associated with birthweight and SSF and valine with cord C-peptide at baseline. The associations of triacylglycerols with birthweight and valine with C-peptide were attenuated when adjusting for maternal BMI or maternal glucose, while the significance of the association between triacylglycerols and SSF was maintained. At fasting, pyruvate demonstrated positive association and acylcarnitine C20 a negative association with SSF, both at baseline and after adjusting for maternal BMI. Glucose adjustment attenuated the association with pyruvate.
At 1 h, several fatty acids, other lipid-related metabolites and amino acids and their metabolites were associated with newborn outcomes across all models. Lauric acid was associated with birthweight at baseline and in model 2. Maternal 1 h triacylglycerols were associated with birthweight, SSF and C-peptide at baseline. The association with birthweight and SSF remained after adjusting for maternal BMI (model 2); however, adjusting for maternal glucose attenuated all associations. NEFA and 3-hydroxybutyrate, a ketone body, were associated with birthweight and C-peptide at baseline and after adjusting for BMI. Additionally, maternal 1 h 3-hydroxybutyrate was associated with SSF at baseline only. One hour 3-hydroxybutyrate and NEFA were not associated with any outcomes after adjusting for maternal glucose. One hour palmitoleic acid, an abundant fatty acid in adipose tissue, was associated with SSF at baseline only. The BCAAs leucine/isoleucine and valine, as well as their carnitine ester, acylcarnitine C4/Ci4, were associated with C-peptide at baseline; however, only valine was associated independent of glucose (model 3) and acylcarnitine C4/Ci4 independent of maternal BMI (model 2). One hour alanine, a gluconeogenic precursor, was associated with birthweight in all models. At 1 h, several other amino acids, including threonine, methionine, ornithine and proline, were associated with birthweight at baseline and after adjusting for maternal BMI or glucose.
Metabolite associations within ancestry groups
Metabolites demonstrating significant associations with newborn outcomes in individual ancestry groups were also observed (ESM Table 3). Fasting pyruvate, 3-indoleacetic acid, threonine and 3-carboxy-4-methyl-5-propyl-2-furanpropanoic acid (CMPF) were associated with birthweight in Afro-Caribbean mother–newborn pairs in all models. At 1 h, there were strong positive associations of leucine/isoleucine, valine, and the BCAA metabolites acylcarnitine C3 and C4/Ci4 with C-peptide in the Afro-Caribbean group in all models (Fig. 2). The associations between fasting CMPF and birthweight in model 1 and 1 h acylcarnitine C4/Ci4 and C-peptide in all models had similar effect sizes in Northern Europeans as in Afro-Caribbeans, but were not significant.
One hour metabolites and newborn outcomes in Afro-Caribbeans. Forest plot depicting 1 h maternal metabolites that are significantly associated with newborn birthweight (a) and cord blood C-peptide (b) in Afro-Caribbean participants. The x-axis represents the standardised β of each individual metabolite. Covariates for each model are as stated in the legend for Fig. 1. Squares, model 1; circles, model 2; diamonds, model 3. AC, acylcarnitine
As shown in Fig. 3, fasting 3-hydroxybutyrate and its carnitine ester acylcarnitine C4-OH were associated with SSF at baseline (model 1) and after adjusting for maternal glucose (model 3) in Northern Europeans, while fasting acylcarnitine C4-OH was also associated with birthweight (model 1) and SSF after adjusting for maternal BMI (model 2).
Fasting metabolites and newborn outcomes in Northern Europeans. Forest plot depicting fasting maternal metabolites that are significantly associated with newborn birthweight (a) and newborn SSF (b) in Northern European participants. The x-axis represents the standardised β of each individual metabolite. Covariates for each model are as stated in the legend for Fig. 1. Squares, model 1; circles, model 2; diamonds, model 3. AC, acylcarnitine
Few ancestry-specific associations were present in Mexican Americans and Thais. Three fasting acylcarnitines (C12-OH/C10-DC, C18:1-OH/C16:1-DC and C5:1) were associated with newborn C-peptide or SSF in Mexican Americans. Fasting triacylglycerols in Thais were associated with SSF in model 1 only. At 1 h in Thais, tyrosine was associated with birthweight in models 1 and 3, while 3-hydroxybutyrate was associated with birthweight in model 1 only. Maternal 1 h 3-hydroxybutyrate and triacylglycerols were associated with SSF in all models, while 1 h leucine/isoleucine and tyrosine were associated with SSF in model 3. The association of fasting and 1 h triacylglycerols with SSF also displayed similar effect sizes in Northern European and Mexican American ancestries, though the associations were not significant.
Network analyses
Recognising that dependencies exist among metabolites, we conducted network analyses to better characterise joint associations of metabolites at both fasting and 1 h with newborn outcomes. Network analyses help identify the context in which metabolite–phenotype associations occur and clarify the interplay between metabolites that do and do not demonstrate individual statistically significant associations with newborn phenotypes.
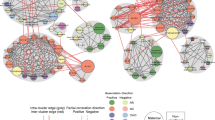
Globally, metabolite networks were larger at 1 h than at fasting but included similar classes of metabolites (Figs. 4, 5, 6, and 7). In maternal metabolite networks associated with newborn C-peptide, several distinct metabolite communities were present. At fasting (Fig. 4), acylcarnitines, amino acids, lipids and fatty acid metabolites were the major metabolites present in models adjusted for maternal BMI (model 2) and maternal glucose (model 3); however, acylcarnitines of all chain lengths were represented in model 3, while only acetylcarnitine C2 and a few medium-chain acylcarnitines were represented in model 2. At 1 h (Fig. 5), several of the same metabolite communities were present in the maternal metabolite networks associated with newborn C-peptide, with the addition of BCAAs and AAAs in both models. In model 2, we also demonstrated the presence of carbohydrate metabolites at 1 h. At both fasting and 1 h, metabolite communities were tightly correlated within metabolite class.
Maternal fasting metabolite subnetworks associated with newborn C-peptide. Subnetwork of maternal fasting metabolites associated with newborn C-peptide in models 2 (a) and 3 (b). Covariates for each model are as stated in the legend for Fig. 1. Blue shading denotes spinglass communities within the estimated networks. The lines between two nodes (edges) represent dependence among metabolite pairs conditional on all other metabolites in the network according to graphical lasso. Solid edges represent dependencies for metabolites in the same spinglass cluster and red dashed edges represent dependencies for metabolites in different spinglass clusters. Large nodes represent metabolites that are individually significant with newborn C-peptide while small nodes are correlated with an individually significant metabolite. Nodes are coloured by metabolite class (amino acid, lipid, glycolysis/tricarboxylic acid cycle, acylcarnitine, fatty acid, miscellaneous, carbohydrate, organic acid, purine/pyrimidine). AA, amino acid; AC, acylcarnitine; Asn/Asx, asparagine/aspartic acid; CHO, carbohydrate; FA, fatty acid; GC/TCA, glycolysis/tricarboxylic acid cycle; G1P, glycerol-1-phosphate; Glu/Glx, glutamine/glutamic acid; Misc, miscellaneous; NM/2AA/NE, N-methylamine/2-aminobutanoic acid/N-ethylglycine; OA, organic acid; 3-OHB, 3-hydroxybutyrate; Pur/Pyr, purine or pyrimidine
Maternal 1 h metabolite subnetworks associated with newborn C-peptide. Subnetwork of maternal 1 h metabolites associated with newborn C-peptide in models 2 (a) and 3 (b). Covariates for each model are as stated in the legend for Fig. 1. Blue shading denotes spinglass communities within the estimated networks. The lines between two nodes (edges) represent dependence among metabolite pairs conditional on all other metabolites in the network according to graphical lasso. Solid edges represent dependencies for metabolites in the same spinglass cluster and red dashed edges represent dependencies for metabolites in different spinglass clusters. Large nodes represent metabolites that are individually significant with newborn C-peptide while small nodes are correlated with an individually significant metabolite. Nodes are coloured by metabolite class (amino acid, lipid, glycolysis/tricarboxylic acid cycle, acylcarnitine, fatty acid, miscellaneous, carbohydrate, organic acid, purine/pyrimidine). AA, amino acid; AC, acylcarnitine; Asn/Asx, asparagine/aspartic acid; CHO, carbohydrate; FA, fatty acid; GC/TCA, glycolysis/tricarboxylic acid cycle; G1P, glycerol-1-phosphate; Glu/Glx, glutamine/glutamic acid; Leu/Ile, leucine/isoleucine; Misc, miscellaneous; NM/2AA/NE, N-methylamine/2-aminobutanoic acid/N-ethylglycine; OA, organic acid; 3-OHB, 3-hydroxybutyrate; OHpro, hydroxyproline; Pur/Pyr, purine or pyrimidine
Maternal fasting metabolite subnetworks associated with newborn SSF. Subnetwork of maternal fasting metabolites associated with newborn SSF in models 2 (a) and 3 (b). Covariates for each model are as stated in the legend for Fig. 1. Blue shading denotes spinglass communities within the estimated networks. The lines between two nodes (edges) represent dependence among metabolite pairs conditional on all other metabolites in the network according to graphical lasso. Solid edges represent dependencies for metabolites in the same spinglass cluster and red dashed edges represent dependencies for metabolites in different spinglass clusters. Large nodes represent metabolites that are individually significant with newborn C-peptide while small nodes are correlated with an individually significant metabolite. Nodes are coloured by metabolite class (amino acid, lipid, glycolysis/tricarboxylic acid cycle, acylcarnitine, fatty acid, miscellaneous, carbohydrate, organic acid, purine/pyrimidine). AA, amino acid; AC, acylcarnitine; Asn/Asx, asparagine/aspartic acid; CHO, carbohydrate; FA, fatty acid; GC/TCA, glycolysis/tricarboxylic acid cycle; G1P, glycerol-1-phosphate; Glu/Glx, glutamine/glutamic acid; Misc, miscellaneous; NM/2AA/NE, N-methylamine/2-aminobutanoic acid/N-ethylglycine; OA, organic acid;3-OHB, 3-hydroxybutyrate; Pur/Pyr, purine or pyrimidine
Maternal 1 h metabolite subnetworks associated with newborn SSF. Subnetwork of maternal 1 h metabolites associated with newborn SSF in models 2 (a) and 3 (b). Covariates for each model are as stated in the legend for Fig. 1. Blue shading denotes spinglass communities within the estimated networks. The lines between two nodes (edges) represent dependence among metabolite pairs conditional on all other metabolites in the network according to graphical lasso. Solid edges represent dependencies for metabolites in the same spinglass cluster and red dashed edges represent dependencies for metabolites in different spinglass clusters. Large nodes represent metabolites that are individually significant with newborn C-peptide while small nodes are correlated with an individually significant metabolite. Nodes are coloured by metabolite class (amino acid, lipid, glycolysis/tricarboxylic acid cycle, acylcarnitine, fatty acid, miscellaneous, carbohydrate, organic acid, purine/pyrimidine). AA, amino acid; AC, acylcarnitine; Asn/Asx, asparagine/aspartic acid; CHO, carbohydrate; FA, fatty acid; GC/TCA, glycolysis/tricarboxylic acid cycle; Glu/Glx, glutamine/glutamic acid; Leu/Ile, leucine/isoleucine; Misc, miscellaneous; NM/2AA/NE, N-methylamine/2-aminobutanoic acid/N-ethylglycine; OA, organic acid; 3-OHB, 3-hydroxybutyrate; OHPro, hydroxyproline; Pur/Pyr, purine or pyrimidine
Maternal fasting metabolites associated with newborn SSF (Fig. 6) included fatty acids, lipid metabolites, amino acids and acylcarnitines in the model adjusted for maternal BMI (model 2). Acylcarnitines formed several clusters, each with acylcarnitines of different chain lengths. Similar classes were represented when adjusting for maternal glucose (model 3); however, the acylcarnitine communities were smaller. At 1 h (Fig. 7), in the models adjusted for maternal BMI or glucose, smaller communities of similar acylcarnitines, fatty acids and lipid metabolites were again represented in the maternal metabolite networks associated with SSF; however, the amino acid community expanded to include all BCAAs and AAAs. In both networks, fatty acids were linked to each other or to lipid metabolites, while amino acids and acylcarnitines were primarily linked to other amino acids and acylcarnitines, respectively. Interconnectivity between clusters occurred primarily between molecules of the same metabolite class; there was minimal connectivity noted between molecules of differing metabolite classes. These data are consistent with different classes of inter-related metabolites contributing to infant body composition independent of maternal glucose and BMI.
Discussion
The current study demonstrates associations of maternal metabolites during pregnancy with newborn adiposity and high cord C-peptide across four ancestry groups in the HAPO cohort. The HAPO study previously demonstrated independent associations of maternal blood glucose levels and BMI with newborn outcomes, including adiposity and C-peptide [11, 20]. Additionally, maternal obesity and gestational diabetes mellitus (GDM) in mothers in the HAPO study had an additive effect on these same newborn outcomes that was greater than either factor alone [21]. The present study suggests that maternal metabolites beyond glucose may contribute to some of these phenotype associations. Several novel metabolite–phenotype associations independent of maternal glucose or BMI were identified, suggesting that additional metabolites contribute to the maternal metabolic milieu and may be reflective of specific maternal exposures, thereby affecting newborn outcomes.
Few existing studies have investigated associations between maternal pregnancy metabolites and newborn size [9, 10] and none have examined maternal metabolite profiles post glucose load or evaluated the relationship between maternal metabolites and newborn adiposity or cord C-peptide. A Spanish cohort of 800 mother–newborn pairs related maternal urinary metabolites during pregnancy with fetal and newborn size and similarly found that maternal urinary BCAAs, alanine, steroid hormones and choline were associated with greater intrauterine fetal growth and birthweight [9]. In a Polish cohort, mothers with lower lipid metabolites and higher adipocyte fatty acid-binding protein at 12–14 weeks of gestation had infants with larger birthweights, hypothesised to be due to greater mother–fetus transport of lipids [10]. However, neither of these studies specifically examined newborn adiposity or insulin resistance and both studied participants with a homogeneous ethnic background.
Our study is the first to identify numerous maternal fatty acid, amino acid and lipid-related metabolites 1 h post glucose load that are associated with newborn birthweight, SSF and C-peptide. Insulin resistance during pregnancy is a normal physiological process that promotes mother–fetus nutrient transfer; this is especially important in the postprandial state. This study and prior studies from our group have identified maternal metabolites associated both with maternal insulin resistance and newborn outcomes, providing insight into which metabolites are important for the regulation of fetal adiposity. Akin to Freinkel’s fuel-mediated teratogenesis hypothesis [22], we propose that maternal metabolic disturbances may have the following consequences: (1) changes in placental transfer of metabolites, including glucose, amino acids and lipid-related metabolites, to the fetus and/or (2) altered development of fetal metabolic tissues in response to exposure to an adverse intrauterine metabolic environment.
We previously demonstrated the association of maternal blood glucose levels and BMI with maternal metabolites [17, 23]; we now show that some of these same metabolites, as well as newly identified metabolites, are associated with newborn size. After adjustment for maternal BMI, maternal fasting glucose was associated with alanine levels; the present study demonstrates that maternal alanine is also associated with birthweight. Maternal 1 h glucose was also associated with 1 h 3-hydroxybutyrate and triacylglycerols, independent of maternal BMI. In the present study, after adjustment for maternal BMI, 1 h 3-hydroxybutyrate levels were associated with birthweight and C-peptide, while 1 h triacylglycerols were associated with birthweight and SSF. Fasting maternal BMI was associated with triacylglycerols independent of glucose levels; in this study, we demonstrate an association of maternal fasting triacylglycerols with birthweight and SSF. The association with birthweight was attenuated when adjusting for maternal BMI or glucose, while the association with SSF was independent of maternal BMI. Thus, maternal glucose elevations may preferentially affect maternal levels of amino acids and ketone bodies, while BMI affects triacylglycerol levels, suggesting that maternal glucose and BMI may contribute to unique maternal metabolic signatures that have an impact on fetal growth and insulin sensitivity.
The positive relationship between maternal triacylglycerols and newborn size independent of maternal BMI in women across the blood glucose spectrum is well described [24]. Though maternal triacylglycerols do not cross the placenta, various placental transporters and receptors facilitate fatty acid transfer to the fetus, providing a substrate for excess fetal growth. While cross-sectional associations between triacylglycerols and newborn size have been established, Mendelian randomisation studies suggest that this association may not be causal and that other factors may contribute [25].
BCAAs and AAAs are related to insulin resistance and type 2 diabetes in adults [6, 26] and obesity and insulin resistance in adolescents [27], although it is unclear whether elevations in these metabolites precede or follow the development of these phenotypes. Some studies in adolescents and adults show that higher BCAA and AAA levels are associated with future insulin resistance and diabetes [27,28,29,30], while more recent studies suggests that BCAA and AAA elevations occur subsequent to the development of adiposity and insulin resistance [31, 32]. Genetic studies have also demonstrated that SNPs associated with elevated BCAA are also strongly associated with type 2 diabetes risk [30, 32]. Although we only identified a relationship between 1 h leucine/isoleucine and birthweight and C-peptide in model 1, 1 h valine was associated with cord C-peptide even after adjustment for maternal BMI or glucose. As maternal BCAAs are a presumed marker of maternal insulin resistance, our findings suggest that elevations in maternal BCAA during pregnancy may relate to newborn size and/or insulin sensitivity. However, we are unable to discern whether this is a direct relationship or indirectly mediated via maternal insulin resistance.
Alanine, a gluconeogenic substrate, has previously been associated with post-load glucose levels in adults [33, 34]. In an insulin-resistant state, insulin-mediated suppression of protein catabolism is reduced; therefore, higher circulating levels of protein-derived gluconeogenic precursors, such as alanine, would be expected. In our cohort, the association of maternal 1 h alanine with newborn birthweight may be a manifestation of higher maternal insulin resistance affecting newborn size, although we did not see a relationship specifically with measures of excess newborn fat. Alternatively, placental transfer of alanine may contribute preferentially to lean body mass development in newborns. 3-Hydroxybutyrate is known to cross the placenta, serving as a substrate for fetal lipogenesis [35]. The associations of maternal 1 h 3-hydroxybutyrate with newborn birthweight and SSF in our cohort suggests that transplacental transfer of 3-hydroxybutyrate may promote growth of fetal adipose tissue, affecting newborn body composition.
Palmitoleic acid, one of the most abundant fatty acids in adipose tissue, is derived primarily from endogenous synthesis and a small amount comes from dietary intake [36]. Palmitoleic acid is readily mobilised from adipose tissue [36] and higher plasma concentrations have been noted in obese children and adults [37, 38]. While some studies have demonstrated a positive correlation between palmitoleic acid levels and diabetes, most show the opposite relationship, suggesting that palmitoleic acid is a measure of insulin sensitivity [36]. In our cohort, 1 h palmitoleic acid was positively associated with SSF, a paradoxical finding in comparison to previous descriptions of the role of this metabolite.
CMPF is a furoic acid that has been implicated in beta cell dysfunction and impaired insulin granule maturation and secretion. CMPF levels have been positively associated with maternal blood glucose levels and GDM and rising levels have been noted over time in non-pregnant individuals that develop type 2 diabetes, due to impaired beta cell function [39,40,41]. In newborns, one would anticipate that markers of maternal insulin resistance, such as CMPF, would be associated with larger newborn size and greater adiposity. In our study, the significant negative association of fasting maternal CMPF with birthweight in Afro-Caribbeans in all models is notable; a positive relationship would have been expected based on previous studies of CMPF reported in the literature.
The network analyses performed in this study highlight the interconnectedness of maternal metabolites as potential contributors to newborn body composition and high cord C-peptide. Acetylcarnitine C2, nearly always the most abundant acylcarnitine in human serum, is a short-chain acylcarnitine thought to be important for the regulation of mitochondrial fuel switching and is elevated in insulin-resistant states [42, 43]. Acylcarnitine C2 was represented in fasting metabolite networks associated with newborn C-peptide and SSF independent of maternal BMI or glucose. Acylcarnitine C2 may be a marker of maternal metabolic inflexibility since it affects the ability of mitochondria to switch between glucose and fatty acid metabolism during a fed or fasted state [44]. When fuel switching is ineffective, mitochondria may become overburdened, leading to acylcarnitine accumulation in tissues; this, in turn, may affect newborn size and insulin action. Interestingly, when examining significant metabolite networks associated with all newborn outcomes at 1 h post glucose when compared with the fasting state, BCAAs and AAAs were consistently represented independent of maternal glucose or BMI. Therefore, these findings in our cohort suggest that the higher levels of these amino acids seen in response to a glucose load are due to maternal insulin resistance limiting insulin-mediated suppression of protein catabolism and may help mediate the well-known association between maternal insulin resistance and newborn size, independent of maternal BMI or glucose levels.
Strengths of this study include its large size and multi-ethnic population, allowing for identification of associations that are shared across ancestries and unique among fewer ancestry groups. Metabolite patterns are influenced by diet and environment and might contribute to the ancestry-specific findings. We also used SSF as a measure of newborn fat mass; this is a more accurate measure than birthweight, weight-for-length or BMI, which do not discriminate between fat mass and fat-free mass [45].
One limitation of this observational study is an inability to establish causality. Evaluating how the association of maternal metabolites and newborn outcomes relates to childhood metabolic disease requires further study in our longitudinal follow-up cohort. Furthermore, metabolites are influenced by many environmental factors, not all of which could be accounted for in the present study.
Maternal metabolites during pregnancy, particularly 1 h post-glucose challenge, are associated with newborn outcomes even when accounting for maternal BMI and glucose. Maternal triacylglycerols, amino acids and acylcarnitines, and their metabolites, may impact fetal growth and adiposity. Further study is needed to delineate the mechanisms underlying these associations.
Data availability
Data will be made available by the authors upon request.
Abbreviations
- AAA:
-
Aromatic amino acid
- BCAA:
-
Branched-chain amino acid
- CMPF:
-
3-Carboxy-4-methyl-5-propyl-2-furanpropanoic acid
- FDR:
-
False discovery rate
- GDM:
-
Gestational diabetes mellitus
- HAPO:
-
Hyperglycemia and Adverse Pregnancy Outcome
- QC:
-
Quality control
- RTL:
-
Retention-time-locking
- SSF:
-
Sum of skinfolds
References
Barker DJ, Osmond C, Simmonds SJ, Wield GA (1993) The relation of small head circumference and thinness at birth to death from cardiovascular disease in adult life. BMJ 306(6875):422–426. https://doi.org/10.1136/bmj.306.6875.422
Gluckman PD, Hanson MA, Mitchell MD (2010) Developmental origins of health and disease: reducing the burden of chronic disease in the next generation. Genome medicine 2(2):14. https://doi.org/10.1186/gm135
Catalano PM, Hauguel-De Mouzon S (2011) Is it time to revisit the Pedersen hypothesis in the face of the obesity epidemic? Am J Obstet Gynecol 204(6):479–487. https://doi.org/10.1016/j.ajog.2010.11.039
Dabelea D, Crume T (2011) Maternal environment and the transgenerational cycle of obesity and diabetes. Diabetes 60(7):1849–1855. https://doi.org/10.2337/db11-0400
Mihalik SJ, Goodpaster BH, Kelley DE et al (2010) Increased levels of plasma acylcarnitines in obesity and type 2 diabetes and identification of a marker of glucolipotoxicity. Obesity (Silver Spring) 18(9):1695–1700. https://doi.org/10.1038/oby.2009.510
Newgard CB, An J, Bain JR et al (2009) A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab 9(4):311–326. https://doi.org/10.1016/j.cmet.2009.02.002
Wang TJ, Larson MG, Vasan RS et al (2011) Metabolite profiles and the risk of developing diabetes. Nat Med 17(4):448–453. https://doi.org/10.1038/nm.2307
Ramos-Roman MA, Sweetman L, Valdez MJ, Parks EJ (2012) Postprandial changes in plasma acylcarnitine concentrations as markers of fatty acid flux in overweight and obesity. Metabolism 61(2):202–212. https://doi.org/10.1016/j.metabol.2011.06.008
Maitre L, Villanueva CM, Lewis MR et al (2016) Maternal urinary metabolic signatures of fetal growth and associated clinical and environmental factors in the INMA study. BMC Med 14(1):177. https://doi.org/10.1186/s12916-016-0706-3
Ciborowski M, Zbucka-Kretowska M, Bomba-Opon D et al (2014) Potential first trimester metabolomic biomarkers of abnormal birth weight in healthy pregnancies. Prenat Diagn 34(9):870–877. https://doi.org/10.1002/pd.4386
HAPO Study Cooperative Research Group (2008) Hyperglycemia and adverse pregnancy outcomes. N Engl J Med 358:1991–2002
Scholtens DM, Bain JR, Reisetter AC et al (2016) Metabolic networks and metabolites underlie associations between maternal glucose during pregnancy and newborn size at birth. Diabetes 65(7):2039–2050. https://doi.org/10.2337/db15-1748
Scholtens DM, Muehlbauer MJ, Daya NR et al (2014) Metabolomics reveals broad-scale metabolic perturbations in hyperglycemic mothers during pregnancy. Diabetes Care 37(1):158–166. https://doi.org/10.2337/dc13-0989
Kind T, Wohlgemuth G, Lee DY et al (2009) FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal Chem 81(24):10038–10048. https://doi.org/10.1021/ac9019522
Halket JM, Przyborowska A, Stein SE, Mallard WG, Down S, Chalmers RA (1999) Deconvolution gas chromatography/mass spectrometry of urinary organic acids--potential for pattern recognition and automated identification of metabolic disorders. Rapid Commun Mass Spectrom 13(4):279–284. https://doi.org/10.1002/(SICI)1097-0231(19990228)13:4<279::AID-RCM478>3.0.CO;2-I
Nodzenski M, Muehlbauer MJ, Bain JR, Reisetter AC, Lowe WL Jr, Scholtens DM (2014) Metabomxtr: an R package for mixture-model analysis of non-targeted metabolomics data. Bioinformatics 30(22):3287–3288. https://doi.org/10.1093/bioinformatics/btu509
Jacob S, Nodzenski M, Reisetter AC et al (2017) Targeted metabolomics demonstrates distinct and overlapping maternal metabolites associated with BMI, glucose, and insulin sensitivity during pregnancy across four ancestry groups. Diabetes Care 40(7):911–919. https://doi.org/10.2337/dc16-2453
Friedman J, Hastie T, Tibshirani R (2008) Sparse inverse covariance estimation with the graphical lasso. Biostatistics (Oxford, England) 9(3):432–441. https://doi.org/10.1093/biostatistics/kxm045
Reichardt J, Bornholdt S (2006) Statistical mechanics of community detection. Phys Rev E Stat Nonlinear Soft Matter Phys 74(1):016110. https://doi.org/10.1103/PhysRevE.74.016110
HAPO Study Cooperative Research Group (2010) Hyperglycaemia and Adverse Pregnancy Outcome (HAPO) Study: associations with maternal body mass index. BJOG 117(5):575–584. https://doi.org/10.1111/j.1471-0528.2009.02486.x
Catalano PM, McIntyre HD, Cruickshank JK et al (2012) The hyperglycemia and adverse pregnancy outcome study: associations of GDM and obesity with pregnancy outcomes. Diabetes Care 35(4):780–786. https://doi.org/10.2337/dc11-1790
Freinkel N (1980) Banting lecture 1980. Of pregnancy and progeny. Diabetes 29(12):1023–1035. https://doi.org/10.2337/diab.29.12.1023
Sandler V, Reisetter AC, Bain JR et al (2017) Associations of maternal BMI and insulin resistance with the maternal metabolome and newborn outcomes. Diabetologia 60(3):518–530. https://doi.org/10.1007/s00125-016-4182-2
Di Cianni G, Miccoli R, Volpe L et al (2005) Maternal triglyceride levels and newborn weight in pregnant women with normal glucose tolerance. Diabet Med 22(1):21–25. https://doi.org/10.1111/j.1464-5491.2004.01336.x
Tyrrell J, Richmond RC, Palmer TM et al (2016) Genetic evidence for causal relationships between maternal obesity-related traits and birth weight. JAMA 315(11):1129–1140. https://doi.org/10.1001/jama.2016.1975
Felig P, Marliss E, Cahill GF Jr (1969) Plasma amino acid levels and insulin secretion in obesity. N Engl J Med 281(15):811–816. https://doi.org/10.1056/NEJM196910092811503
McCormack SE, Shaham O, McCarthy MA et al (2013) Circulating branched-chain amino acid concentrations are associated with obesity and future insulin resistance in children and adolescents. Pediatr Obes 8(1):52–61. https://doi.org/10.1111/j.2047-6310.2012.00087.x
Wurtz P, Soininen P, Kangas AJ et al (2013) Branched-chain and aromatic amino acids are predictors of insulin resistance in young adults. Diabetes Care 36(3):648–655. https://doi.org/10.2337/dc12-0895
Chen T, Ni Y, Ma X et al (2016) Branched-chain and aromatic amino acid profiles and diabetes risk in Chinese populations. Sci Rep 6(1):20594. https://doi.org/10.1038/srep20594
Lotta LA, Scott RA, Sharp SJ et al (2016) Genetic predisposition to an impaired metabolism of the branched-chain amino acids and risk of type 2 diabetes: a Mendelian randomisation analysis. Plos Med 13(11):e1002179. https://doi.org/10.1371/journal.pmed.1002179
Mahendran Y, Jonsson A, Have CT et al (2017) Genetic evidence of a causal effect of insulin resistance on branched-chain amino acid levels. Diabetologia 60(5):873–878. https://doi.org/10.1007/s00125-017-4222-6
Wang Q, Holmes MV, Davey Smith G, Ala-Korpela M (2017) Genetic support for a causal role of insulin resistance on circulating branched-chain amino acids and inflammation. 40(12):1779–1786. https://doi.org/10.2337/dc17-1642
Wurtz P, Tiainen M, Makinen VP et al (2012) Circulating metabolite predictors of glycemia in middle-aged men and women. Diabetes Care 35(8):1749–1756. https://doi.org/10.2337/dc11-1838
Guasch-Ferre M, Hruby A, Toledo E, Clish CB, Martinez-Gonzalez MA, Salas-Salvado J (2016) Metabolomics in prediabetes and diabetes: a systematic review and meta-analysis. Diabetes Care 39(5):833–846. https://doi.org/10.2337/dc15-2251
Herrera E, Amusquivar E, Lopez-Soldado I, Ortega H (2006) Maternal lipid metabolism and placental lipid transfer. Horm Res 65 Suppl 3:59–64. https://doi.org/10.1159/000091507
Frigolet ME, Gutierrez-Aguilar R (2017) The role of the novel lipokine palmitoleic acid in health and disease. Adv Nutr (Bethesda, Md) 8(1):173s–181s. https://doi.org/10.3945/an.115.011130
Okada T, Furuhashi N, Kuromori Y, Miyashita M, Iwata F, Harada K (2005) Plasma palmitoleic acid content and obesity in children. Am J Clin Nutr 82(4):747–750. https://doi.org/10.1093/ajcn/82.4.747
Paillard F, Catheline D, Duff FL et al (2008) Plasma palmitoleic acid, a product of stearoyl-coA desaturase activity, is an independent marker of triglyceridemia and abdominal adiposity. Nutr Metab Cardiovasc Dis 18(6):436–440. https://doi.org/10.1016/j.numecd.2007.02.017
Liu Y, Prentice KJ, Eversley JA et al (2016) Rapid elevation in CMPF may act as a tipping point in diabetes development. Cell Rep 14(12):2889–2900. https://doi.org/10.1016/j.celrep.2016.02.079
Prentice KJ, Luu L, Allister EM et al (2014) The furan fatty acid metabolite CMPF is elevated in diabetes and induces beta cell dysfunction. Cell Metab 19(4):653–666. https://doi.org/10.1016/j.cmet.2014.03.008
Retnakaran R, Ye C, Kramer CK et al (2016) Evaluation of circulating determinants of beta-cell function in women with and without gestational diabetes. J Clin Endocrinol Metab 101(7):2683–2691. https://doi.org/10.1210/jc.2016-1402
Schooneman MG, Vaz FM, Houten SM, Soeters MR (2013) Acylcarnitines: reflecting or inflicting insulin resistance? Diabetes 62(1):1–8. https://doi.org/10.2337/db12-0466
Muoio DM, Noland RC, Kovalik JP et al (2012) Muscle-specific deletion of carnitine acetyltransferase compromises glucose tolerance and metabolic flexibility. Cell Metab 15(5):764–777. https://doi.org/10.1016/j.cmet.2012.04.005
Muoio DM (2014) Metabolic inflexibility: when mitochondrial indecision leads to metabolic gridlock. Cell 159(6):1253–1262. https://doi.org/10.1016/j.cell.2014.11.034
Chen LW, Tint MT, Fortier MV et al (2017) Which anthropometric measures best reflect neonatal adiposity? Int J Obes 42(3):501–506. https://doi.org/10.1038/ijo.2017.250
Funding
This study was funded by grants from the National Institute of Diabetes and Digestive and Kidney Diseases (R01DK095963) and the National Institute of Child Health and Human Development (R01-HD34242, R01-HD34243).
Author information
Authors and Affiliations
Consortia
Contributions
RK contributed to data interpretation and manuscript writing. MN, OT and AK contributed to analysis and interpretation of data. MN and DMS led data analysis. JRB, MJM, RDS, ORI, SON, LPL, BEM and CBN contributed to acquisition and interpretation of data. DMS and WLL were involved in all aspects of the study, including study design and data collection, analysis and interpretation. All authors made critical intellectual contributions to drafting and/or revising the manuscript and all approved the final version. DMS and WLL are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Corresponding author
Ethics declarations
The authors declare that there is no duality of interest associated with this manuscript.
Electronic supplementary material
Tables
(PDF 181 kb)
Rights and permissions
About this article
Cite this article
Kadakia, R., Nodzenski, M., Talbot, O. et al. Maternal metabolites during pregnancy are associated with newborn outcomes and hyperinsulinaemia across ancestries. Diabetologia 62, 473–484 (2019). https://doi.org/10.1007/s00125-018-4781-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-018-4781-1