Abstract
Key message
A novel genomic region controlling thermotolerance at flowering was identified by the combination of whole genomic re-sequencing and bulked segregant analysis in maize.
Abstract
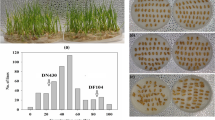
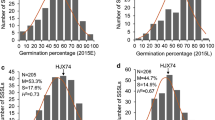
The increasing frequency of extreme high temperature has brought a great threat to the development of maize throughout its life cycle, especially during the flowering phase. However, the genetic basis of thermotolerance at flowering in maize remains poorly understood. Here, we characterized a thermotolerant maize ecotype Abe2 and dissected its genetic basis using a F2:8 recombinant inbred line (RIL) population generated from a cross between Abe2 and B73. After continuous high temperature stress above 35 °C for 17 days, Abe2 and B73 show distinct leaf scorching phenotype under field conditions. To identify the genomic regions associated with the phenotypic variation, we applied a combination of whole genomic re-sequencing and bulked segregant analysis, and revealed 10,316,744 SNPs and 1,488,302 InDels between the two parental lines, and 2,693,054 SNPs and 313,757 InDels between the two DNA pools generated from the thermos-tolerant and the sensitive individuals of the RIL, of which, 108,655 and 17,853 SNPs may cause nonsynonymous variations. Finally, a 7.41 Mb genomic region on chromosome 1 was identified, and 7 candidate genes were annotated to participate in high temperature-related stress response. A candidate gene Zm00001d033339 encoding a serine/threonine protein kinase was proposed to be the most likely causative gene contributing to the thermotolerance at flowering by involving in stomatal movement (GO: 0010119) via Abscisic acid (ABA) pathway (KO04075). This work could provide an opportunity for gene cloning and pyramiding breeding to improve thermotolerance at flowering in maize.





Similar content being viewed by others
Data availability
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive (Wang et al. 2017) in BIG Data Center (BIG et al. 2019), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession numbers CRA002322, CRA002322 that are publicly accessible at https://bigd.big.ac.cn/gsa. The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
References
Acharya BR, Jeon BW, Zhang W, Assmann SM (2013) Open Stomata 1 (OST1) is limiting in abscisic acid responses of Arabidopsis guard cells. New Phytol 200:1049–1063. https://doi.org/10.1111/nph.12469
Balfagon D, Zandalinas SI, Gomezcadenas A (2019) High temperatures change the perspective: integrating hormonal responses in citrus plants under co-occurring abiotic stress conditions. Physiol Plant 165:183–197. https://doi.org/10.1111/ppl.12815
BIG Data Center Members (2019) Database resources of the BIG Data Center in 2019. Nucleic Acids Res 47:D8–D14. https://doi.org/10.1093/nar/gky993
Bita CE, Gerats T (2013) Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Front Plant Sci 4:273. https://doi.org/10.3389/fpls.2013.00273
Brugiere N, Zhang W, Xu J, Scolaro E, Lu C, Kahsay RY, Kise R, Trecker L, Williams RW, Hakimi SM (2017) Overexpression of RING domain E3 ligase ZmXerico1 confers drought tolerance through regulation of ABA homeostasis. Plant Physiol 175:1350–1369. https://doi.org/10.1104/pp.17.01072
Burghardt LT, Runcie DE, Wilczek AM, Cooper MD, Roe JL, Welch SM, Schmitt J (2016) Fluctuating, warm temperatures decrease the effect of a key floral repressor on flowering time in Arabidopsis thaliana. New Phytol 210:564–576. https://doi.org/10.1111/nph.13799
Cao Y, Han Y, Jin Q, Lin Y, Cai Y (2016a) Comparative genomic analysis of the GRF genes in Chinese pear (Pyrus bretschneideri Rehd), poplar (Populous), grape (Vitis vinifera), Arabidopsis and rice (Oryza sativa). Front Plant Sci 7:1750. https://doi.org/10.3389/fpls.2016.01750
Cao Y, Han Y, Li D, Lin Y, Cai Y (2016b) MYB transcription factors in Chinese pear (Pyrus bretschneideri Rehd.): genome-wide identification, classification, and expression profiling during fruit development. Front Plant Sci 7:577. https://doi.org/10.3389/fpls.2016.00577
Cao Y, Han Y, Meng D, Abdullah M, Li D, Jin Q, Lin Y, Cai Y (2018) Systematic analysis and comparison of the PHD—finger gene family in Chinese pear (Pyrus bretschneideri) and its role in fruit development. Funct Integr Genomics 18:519–531. https://doi.org/10.1007/s10142-018-0609-9
Cingolani P, Platts A, Le Wang L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92. https://doi.org/10.4161/fly.19695
Dupuis I, Dumas C (1990) Influence of temperature stress on in vitro fertilization and heat shock protein synthesis in maize (Zea mays L.) reproductive tissues. Plant Physiol 94:665–670. https://doi.org/10.1104/pp.94.2.665
Fahad S, Bajwa AA, Nazir U, Anjum SA, Farooq A, Zohaib A, Sadia S, Nasim W, Adkins SW, Saud S (2017) Crop production under drought and heat stress: plant responses and management options. Front Plant Sci 8:1147. https://doi.org/10.3389/fpls.2017.01147
Fekih R, Takagi H, Tamiru M, Abe A, Natsume S, Yaegashi H, Sharma S, Sharma S, Kanzaki H, Matsumura H (2013) MutMap+ : genetic mapping and mutant identification without crossing in rice. PLoS ONE. https://doi.org/10.1371/journal.pone.0068529
Frey FP, Presterl T, Lecoq P, Orlik A, Stich B (2016) First steps to understand heat tolerance of temperate maize at adult stage: identification of QTL across multiple environments with connected segregating populations. Theor Appl Genet 129:945–961. https://doi.org/10.1007/s00122-016-2674-6
Gao J, Wang S, Zhou Z, Wang S, Dong C, Mu C, Song Y, Ma P, Li C, Wang Z, He K, Han C, Chen J, Yu H, Wu J (2019) Linkage mapping and genome-wide association reveal candidate genes conferring thermotolerance of seed-set in maize. J Exp Bot. https://doi.org/10.1093/jxb/erz171
Gourdji SM, Sibley AM, Lobell DB (2013) Global crop exposure to critical high temperatures in the reproductive period: historical trends and future projections. Environ Res Lett 8:024041. https://doi.org/10.1088/1748-9326/8/2/024041
Hatfield JL, Prueger JH (2015) Temperature extremes: effect on plant growth and development. Weather Clim Extremes 10:4–10. https://doi.org/10.1093/ee/nvx111
Hill JT, Demarest BL, Bisgrove BW, Gorsi B, Su YC, Yost HJ (2013) MMAPPR: mutation mapping analysis pipeline for pooled RNA-seq. Genome Res 23:687–697. https://doi.org/10.1101/gr.146936.112
Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32:277–280. https://doi.org/10.1093/nar/gkh063
Kim S, Kang J, Cho D, Park JH, Kim SY (2004) ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J 40:75–87. https://doi.org/10.1111/j.1365-313x.2004.02192.x
Lahari Z, Ribeiro A, Talukdar P, Martin B, Heidari Z, Gheysen G, Price AH, Shrestha R (2019) QTL-seq reveals a major root-knot nematode resistance locus on chromosome 11 in rice (Oryza sativa L.). Euphytica 215:117. https://doi.org/10.1007/s10681-019-2427-0
Larkindale J, Hall JD, Knight MR, Vierling E (2005) Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol 138:882–897. https://doi.org/10.1104/pp.105.062257
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp698
Liu C, Zhou Q, Dong LS, Wang HM, Liu F, Weng J, Li X, Xie C (2016) Genetic architecture of the maize kernel row number revealed by combining QTL mapping using a high-density genetic map and bulked segregant RNA sequencing. BMC Genomics 17:915. https://doi.org/10.1186/s12864-016-3240-y
Lizaso JI, Ruizramos M, Rodriguez L, Gabaldonleal C, Oliveira JA, Lorite IJ, Sanchez D, Garcia E, Rodriguez A (2018) Impact of high temperatures in maize: phenology and yield components. Field Crops Res 216:129–140. https://doi.org/10.1016/j.fcr.2017.11.013
Lu H, Lin T, Klein J, Wang S, Qi J, Zhou Q, Sun J, Zhang Z, Weng Y, Huang S (2014) QTL-seq identifies an early flowering QTL located near Flowering Locus T in cucumber. Theor Appl Genet 127:1491–1499. https://doi.org/10.1007/s00122-014-2313-z
Mckenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky AM, Garimella K, Altshuler D, Gabriel S, Daly MJ (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. https://doi.org/10.1101/gr.107524.110
Mittler R, Finka A, Goloubinoff P (2012) How do plants feel the heat. Trends Biochem Sci 37:118–125. https://doi.org/10.1016/j.tibs.2011.11.007
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325. https://doi.org/10.1093/nar/8.19.4321
Nakamura K, Oshima T, Morimoto T, Ikeda S, Yoshikawa H, Shiwa Y, Ishikawa S, Linak MC, Hirai A, Takahashi H, Altaf-Ul-Amin M, Ogasawara N, Kanaya S (2011) Sequence-specific error profile of illumina sequencers. Nucleic Acids Res 39:e90. https://doi.org/10.1093/nar/gkr344
Nakano Y, Higuchi Y, Sumitomo K, Hisamatsu T (2013) Flowering retardation by high temperature in chrysanthemums: involvement of FLOWERING LOCUS T-like 3 gene repression. J Exp Bot 64:909–920. https://doi.org/10.1093/jxb/ers370
Ning H (2012) Field trials and statistical methods (in Chinese). In: Ning H (ed) Common test design, 1st edn. Science Press, Beijing, pp 14–17
Obata T, Witt S, Lisec J, Palaciosrojas N, Florezsarasa I, Yousfi S, Araus JL, Cairns JE, Fernie AR (2015) Metabolite profiles of maize leaves in drought, heat, and combined stress field trials reveal the relationship between metabolism and grain yield. Plant Physiol 169:2665–2683. https://doi.org/10.1104/pp.15.01164
Pang W, Fu P, Li X, Zhan Z, Yu S, Piao Z (2018) Identification and mapping of the clubroot resistance gene CRd in Chinese cabbage (Brassica rapa ssp. pekinensis). Front Plant Sci 9:653. https://doi.org/10.3389/fpls.2018.00653
Qian Y, Hu W, Liao J, Zhang J, Ren Q (2019a) The dynamics of DNA methylation in the maize (Zea mays L.) inbred line B73 response to heat stress at the seedling stage. Biochem Biophys Res Commun 512:742–749. https://doi.org/10.1016/j.bbrc.2019.03.150
Qian Y, Ren Q, Zhang J, Chen L (2019b) Transcriptomic analysis of the maize (Zea mays L.) inbred line B73 response to heat stress at the seedling stage. Gene 692:68–78. https://doi.org/10.1016/j.gene.2018.12.062
Samakovli D, Tichá T, Vavrdová T, Ovečka M, Luptovčiak I, Zapletalová V, Kuchařová A, Křenek P, Krasylenko Y, Margaritopoulou T, Roka L, Milioni D, Komis G, Hatzopoulos P, Šamaj J (2020) YODA-HSP90 module regulates phosphorylation-dependent inactivation of SPEECHLESS to control stomatal development under acute heat stress in Arabidopsis. Mol Plant. https://doi.org/10.1016/j.molp.2020.01.001
Schoper J, Lambert RJ, Vasilas BL, Westgate ME (1987) Plant factors controlling seed set in maize the influence of silk, pollen, and ear-leaf water status and tassel heat treatment at pollination. Plant Physiol 83:121–125. https://doi.org/10.1104/pp.83.1.121
Semenov MA, Stratonovitch P, Alghabari F, Gooding MJ (2014) Adapting wheat in Europe for climate change. J Cereal Sci 59:245–256. https://doi.org/10.1016/j.jcs.2014.01.006
Shen H, Zhong X, Zhao F, Wang Y, Yan B, Li Q, Chen G, Mao B, Wang J, Li Y, Xiao G, He Y, Xiao H, Li J, He Z (2015) Overexpression of receptor-like kinase ERECTA improves thermotolerance in rice and tomato. Nat Biotechnol 33:996–1003. https://doi.org/10.1038/nbt.3321
Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S, Mitsuoka C, Uemura A, Utsushi H, Tamiru M, Takuno S (2013) QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J 74:174–183. https://doi.org/10.1111/tpj.12105
Tatusov RL, Galperin MY, Natale DA, Koonin EV (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28(1):33–36. https://doi.org/10.1093/nar/28.1.33
Tito R, Vasconcelos HL, Feeley KJ (2018) Global climate change increases risk of crop yield losses and food insecurity in the tropical Andes. Glob Change Biol 24(2):e592–e602. https://doi.org/10.1111/gcb.13959
Wang P, Zhao L, Hou H, Zhang H, Huang Y, Wang Y, Li H, Gao F, Yan S, Li L (2015) Epigenetic changes are associated with programmed cell death induced by heat stress in seedling leaves of Zea mays. Plant Cell Physiol 56:965–976. https://doi.org/10.1093/pcp/pcv023
Wang Y, Song F, Zhu J, Zhang S, Yang Y, Chen T, Tang B, Dong L, Ding N, Zhang Q, Bai Z, Dong X, Chen H, Sun M, Zhai S, Sun Y, Yu L, Lan L, Xiao J, Fang X, Lei H, Zhang Z, Zhao W (2017) GSA: genome sequence archive. Genomics Proteomics Bioinform 15(1):14–18. https://doi.org/10.1016/j.gpb.2017.01.001
Watson M (2014) Quality assessment and control of high-throughput sequencing data. Front Genet 5:235. https://doi.org/10.3389/fgene.2014.00235
Wu J, Wang L, Lin Y, Weng C, Yeh C, Wu S (2017) The Arabidopsis heat-intolerant 5 (hit5)/enhanced response to aba 1 (era1) mutant reveals the crucial role of protein farnesylation in plant responses to heat stress. New Phytol 213:1181–1193. https://doi.org/10.1111/nph.14212
Xu Y, Ramanathan V, Victor DG (2018) Global warming will happen faster than we think. Nature 564:30–32. https://doi.org/10.1038/d41586-018-07586-5
Yamamoto K, Sakamoto H, Momonoki YS (2011) Maize acetylcholinesterase is a positive regulator of heat tolerance in plants. J Plant Physiol 168:1987–1992. https://doi.org/10.1016/j.jplph.2011.06.001
Zandalinas SI, Balfagon D, Arbona V, Gomezcadenas A, Inupakutika MA, Mittler R (2016) ABA is required for the accumulation of APX1 and MBF1c during a combination of water deficit and heat stress. J Exp Bot 67:5381–5390. https://doi.org/10.1093/jxb/erw299
Zhao F, Zhang D, Zhao Y, Wang W, Yang H, Tai F, Li C, Hu X (2016) The difference of physiological and proteomic changes in maize leaves adaptation to drought, heat, and combined both stresses. Front Plant Sci 7:1471. https://doi.org/10.3389/fpls.2016.01471
Zhong C, Sun S, Li Y, Duan C, Zhu Z (2018) Next-generation sequencing to identify candidate genes and develop diagnostic markers for a novel Phytophthora resistance gene, RpsHC18, in soybean. Theor Appl Genet 131:525–538. https://doi.org/10.1007/s00122-017-3016-z
Zhu P, Zhuang Q, Archontoulis S, Bernacchi CJ, Muller C (2019a) Dissecting the nonlinear response of maize yield to high temperature stress with model-data integration. Glob Change Biol 25(7):2470–2484. https://doi.org/10.1111/gcb.14632
Zhu X, Gou X, Zhou M (2019b) Nomograms predict survival advantages of Gleason score 3 + 4 over 4 + 3 for prostate cancer: a SEER-based study. Front Oncol 9:646. https://doi.org/10.3389/fonc.2019.00646
Acknowledgements
We thank Dr. Chuanxiao Xie from the Institute of Crop Science, Chinese Academy of Agricultural Sciences for providing the maize ecotype Abe2. This work was supported by The National Key Research and Development Program of China (2016YFD0101803), the National Key Research and Development Program of China (2017YFD0301301) and the National Natural Science Foundation of China (31670264). No conflict of interest declared.
Author information
Authors and Affiliations
Contributions
PL, WZ, JS and CQ contributed conception and designed the study. WZ, JS, CQ, SY, SH, CH, YW, WW, HC, SR, CC, ZT and CW performed the field experiments. WZ assayed the plant phenotype under controlled growth conditions, assembled and analyzed the data. WZ and PL wrote the manuscript. All authors contributed to manuscript revision.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Antonio Augusto Franco Garcia.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zeng, W., Shi, J., Qiu, C. et al. Identification of a genomic region controlling thermotolerance at flowering in maize using a combination of whole genomic re-sequencing and bulked segregant analysis. Theor Appl Genet 133, 2797–2810 (2020). https://doi.org/10.1007/s00122-020-03632-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-020-03632-x