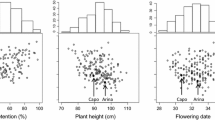
Abstract
Key message
QTL analyses of two bi-parental mapping populations with AC Barrie as a parent revealed numerous FHB-resistance QTL unique to each population and uncovered novel variation near Fhb1.
Abstract
Fusarium head blight (FHB) is a destructive disease of wheat worldwide, leading to severe yield and quality losses. The genetic basis of native FHB resistance was examined in two populations: a recombinant inbred line population from the cross Cutler/AC Barrie and a doubled haploid (DH) population from the cross AC Barrie/Reeder. Numerous QTL were detected among the two mapping populations with many being cross-specific. Photoperiod insensitivity at Ppd-D1 and dwarfing at Rht-B1 and Rht-D1 was associated with increased FHB susceptibility. Anthesis date QTL at or near the Vrn-A1 and Vrn-B1 loci co-located with major FHB-resistance QTL in the AC Barrie/Reeder population. The loci were epistatic for both traits, such that DH lines with both late alleles were considerably later to anthesis and had reduced FHB symptoms (i.e., responsible for the epistatic interaction). Interestingly, AC Barrie contributed FHB resistance near the Fhb1 locus in the Cutler population and susceptibility in the Reeder population. Analyses of the Fhb1 candidate genes PFT and TaHRC confirmed that AC Barrie, Cutler, and Reeder do not carry the Sumai-3 Fhb1 gene. Resistance QTL were also detected at the expected locations of Fhb2 and Fhb5. The native FHB-resistance QTL detected near Fhb1, Fhb2, and Fhb5 do not appear to be as effective as Fhb1, Fhb2, and Fhb5 from Sumai-3. The presence of awns segregated at the B1 awn inhibitor locus in both populations, but was only associated with FHB resistance in the Cutler/AC Barrie population suggesting linkage caused the association rather than pleiotropy.



Similar content being viewed by others
Availability of data and material
Data supporting the current study can be obtained by contacting the corresponding author (curt.mccartney@canada.ca).
Abbreviations
- ANOVA:
-
Analysis of variance
- BLAST:
-
Basic local alignment search tool
- BLUPs:
-
Best linear unbiased predictors
- C/B:
-
Cutler/AC Barrie
- DH:
-
Doubled haploid
- ICIM:
-
Inclusive composite interval mapping
- IM:
-
Interval mapping
- LOD:
-
Logarithm of odds
- META-R:
-
Multi-environment trial analysis with R
- PVE:
-
Phenotypic variation explained
- QTL:
-
Quantitative trait loci
- B/R:
-
AC Barrie/Reeder
- RIL:
-
Recombinant inbred line
- SNP:
-
Single-nucleotide polymorphism
References
Alvarado G, López M, Vargas M, Pacheco Á, Rodríguez F, Burgueño J, Crossa J (2017) META-R (Multi Environment Trail Analysis with R for Windows) Version 6.01. CIMMYT Research Data & Software Repository Network. https://hdl.handle.net/11529/10201
Appels R, Eversole K, Feuillet C, Keller B, Rogers J, Stein N, Pozniak CJ, Stein N, Choulet F, Distelfeld A et al (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. https://doi.org/10.1126/science.aar7191
Bai G, Shaner G (2004) Management and resistance in wheat and barley to Fusarium head blight. Annu Rev Phytopathol 42:135–161. https://doi.org/10.1146/annurev.phyto.42.040803.140340
Bai G, Su Z, Cai J (2018) Wheat resistance to Fusarium head blight. Can J Plant Pathol 40:336–346. https://doi.org/10.1080/07060661.2018.1476411
Beales J, Turner A, Griffiths S, Snape JW, Laurie DA (2007) A Pseudo-Response Regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor Appl Genet 115:721–733. https://doi.org/10.1007/s00122-007-0603-4
Blake NK, Lanning SP, Berg JE, Bruckner PL, Sherman JD, Talbert LE (2011) Registration of spring- and winter-habit wheat lines derived from elite cultivars of the alternate growth habit. J Plant Registr 5:418–421. https://doi.org/10.3198/jpr2011.01.0003crg
Blake NK, Lanning SP, Martin JM, Doyle M, Sherman JD, Naruoka Y, Talbert LE (2009) Effect of variation for major growth habit genes on maturity and yield in five spring wheat populations. Crop Sci 49:1211–1220. https://doi.org/10.2135/cropsci2008.08.0505
Bonin CM, Kolb FL (2009) Resistance to fusarium head blight and kernel damage in a winter wheat recombinant inbred line population. Crop Sci 49:1304–1312. https://doi.org/10.2135/cropsci2008.08.0459
Briggs KG, Kibite S, Kutschera K (1992) Cutler red spring wheat. Can J Plant Sci 72:229–233
Buerstmayr H, Ban T, Anderson JA (2009) QTL mapping and marker-assisted selection for Fusarium head blight resistance in wheat: a review. Plant Breed 128:1–26. https://doi.org/10.1111/j.1439-0523.2008.01550.x
Buerstmayr H, Lemmens M, Hartl L, Doldi L, Steiner B, Stierschneider M, Ruckenbauer P (2002) Molecular mapping of QTLs for Fusarium head blight resistance in spring wheat. I. Resistance to fungal spread (type II resistance). Theor Appl Genet 104:84–91. https://doi.org/10.1007/s001220200009
Buerstmayr M, Huber K, Heckmann J, Steiner B, Nelson JC, Buerstmayr H (2012) Mapping of QTL for Fusarium head blight resistance and morphological and developmental traits in three backcross populations derived from Triticum dicoccum x Triticum durum. Theor Appl Genet 125:1751–1765. https://doi.org/10.1007/s00122-012-1951-2
Buerstmayr M, Steiner B, Wagner C, Schwarz P, Brugger K, Barabaschi D, Volante A, Valè G, Cattivelli L, Buerstmayr H (2018) High-resolution mapping of the pericentromeric region on wheat chromosome arm 5AS harbouring the Fusarium head blight resistance QTL Qfhs.ifa-5A. Plant Biotechnol J 16:1046–1056. https://doi.org/10.1111/pbi.12850
Canadian Grain Commission (2019) Frequency and severity of Fusarium damaged kernels (FDK) in Harvest Sample Program red spring wheat samples. https://www.grainscanada.gc.ca/en/grain-research/export-quality/cereals/wheat/western/annual-fusarium-damage/canada-western-red-spring/. Accessed 13 Dec 2019
Cavanagh CR, Chao S, Wang S, Huang BE, Stephen S, Kiani S, Forrest K, Saintenac C, Brown-Guedira GL, Akhunova A, See D, Bai G, Pumphrey M, Tomar L, Wong D, Kong S, Reynolds M, da Silva ML, Bockelman H, Talbert L, Anderson JA, Dreisigacker S, Baenziger S, Carter A, Korzun V, Morrell PL, Dubcovsky J, Morell MK, Sorrells ME, Hayden MJ, Akhunov E (2013) Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. P Natl Acad Sci USA 110:8057–8062. https://doi.org/10.1073/pnas.1217133110
Chen H, Moakhar NP, Iqbal M, Pozniak C, Hucl P, Spaner D (2016) Genetic variation for flowering time and height reducing genes and important traits in western Canadian spring wheat. Euphytica 208:377–390. https://doi.org/10.1007/s10681-015-1615-9
Cuthbert PA, Somers DJ, Brule-Babel A (2007) Mapping of Fhb2 on chromosome 6BS: a gene controlling Fusarium head blight field resistance in bread wheat (Triticum aestivum L.). Theor Appl Genet 114:429–437. https://doi.org/10.1007/s00122-006-0439-3
Cuthbert PA, Somers DJ, Thomas J, Cloutier S, Brulé-Babel A (2006) Fine mapping Fhb1, a major gene controlling fusarium head blight resistance in bread wheat (Triticum aestivum L.). Theor Appl Genet 112:1465–1472. https://doi.org/10.1007/s00122-006-0249-7
Draeger R, Gosman N, Steed A, Chandler E, Thomsett M, Srinivasachary SJ, Buerstmayr H, Lemmens M, Schmolke M, Mesterhazy A, Nicholson P (2007) Identification of QTLs for resistance to Fusarium head blight, DON accumulation and associated traits in the winter wheat variety Arina. Theor Appl Genet 115:617–625. https://doi.org/10.1007/s00122-007-0592-3
Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred I Accuracy assessment. Genome Res 8:175–185
Ferrigo D, Raiola A, Causin R (2016) Fusarium toxins in cereals: Occurrence, legislation, factors promoting the appearance and their management. Molecules. https://doi.org/10.3390/molecules21050627
Giancaspro A, Giove SL, Zito D, Blanco A, Gadaleta A (2016) Mapping QTLs for Fusarium head blight resistance in an interspecific wheat population. Front Plant Sci 7:1381. https://doi.org/10.3389/fpls.2016.01381
Gilbert J, Haber S (2013) Overview of some recent research developments in Fusarium head blight of wheat. Can J Plant Pathol 35:149–174. https://doi.org/10.1080/07060661.2013.772921
Gilbert J, Tekauz A (2000) Review: Recent developments in research on Fusarium head blight of wheat in Canada. Can J Plant Pathol 22:1–8. https://doi.org/10.1080/07060660009501155
Handa H, Namiki N, Xu D, Ban T (2008) Dissecting of the FHB resistance QTL on the short arm of wheat chromosome 2D using a comparative genomic approach: from QTL to candidate gene. Mol Breed 22:71–84. https://doi.org/10.1007/s11032-008-9157-7
He Y, Zhang X, Zhang Y, Ahmad D, Wu L, Jiang P, Ma H (2018) Molecular characterization and expression of PFT, an FHB resistance gene at the Fhb1 QTL in wheat. Phytopathology 108:730–736. https://doi.org/10.1094/PHYTO-11-17-0383-R
Holzapfel J, Voss HH, Miedaner T, Korzun V, Häberle J, Schweizer G, Mohler V, Zimmermann G, Hartl L (2008) Inheritance of resistance to Fusarium head blight in three European winter wheat populations. Theor Appl Genet 117:1119–1128. https://doi.org/10.1007/s00122-008-0850-z
Hu X, Rocheleau H, McCartney C, Biselli C, Bagnaresi P, Balcerzak M, Fedak G, Yan Z, Valè G, Khanizadeh S, Ouellet T (2019) Identification and mapping of expressed genes associated with the 2DL QTL for fusarium head blight resistance in the wheat line Wuhan 1. BMC Genet 20:47. https://doi.org/10.1186/s12863-019-0748-6
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877
Iqbal M, Navabi A, Salmon DF, Yang RC, Spaner D (2006) A genetic examination of early flowering and maturity in Canadian spring wheat. Can J Plant Sci 86:995–1004. https://doi.org/10.4141/P06-002
Kosambi DD (1943) The estimation of map distances from recombination values. Ann Eugenic 12:172–175. https://doi.org/10.1111/j.1469-1809.1943.tb02321.x
Li G, Zhou J, Jia H, Gao Z, Fan M, Luo Y, Zhao P, Xue S, Li N, Yuan Y, Ma S, Kong Z, Jia L, An X, Jiang G, Liu W, Cao W, Zhang R, Fan J, Xu X, Liu Y, Kong Q, Zheng S, Wang Y, Qin B, Cao S, Ding Y, Shi J, Yan H, Wang X, Ran C, Ma Z (2019) Mutation of a histidine-rich calcium-binding-protein gene in wheat confers resistance to Fusarium head blight. Nat Genet. https://doi.org/10.1038/s41588-019-0426-7
Li H, Ribaut JM, Li Z, Wang J (2008) Inclusive composite interval mapping (ICIM) for digenic epistasis of quantitative traits in biparental populations. Theor Appl Genet 116:243–260. https://doi.org/10.1007/s00122-007-0663-5
Li H, Ye G, Wang J (2007) A modified algorithm for the improvement of composite interval mapping. Genetics 175:361–374. https://doi.org/10.1534/genetics.106.066811
Liu S, Hall MD, Griffey CA, McKendry AL (2009) Meta-Analysis of QTL associated with Fusarium head blight resistance in wheat. Crop Sci 49:1955–1968. https://doi.org/10.2135/cropsci2009.03.0115
Liu S, Pumphrey M, Gill B, Trick H, Zhang J, Dolezel J, Chalhoub B, Anderson J (2008) Toward positional cloning of Fhb1, a major QTL for Fusarium head blight resistance in wheat. Cereal Res Commun 36:195–201. https://doi.org/10.1556/CRC.36.2008.Suppl.B.15
Liu S, Zhang X, Pumphrey MO, Stack RW, Gill BS, Anderson JA (2006) Complex microcolinearity among wheat, rice, and barley revealed by fine mapping of the genomic region harboring a major QTL for resistance to Fusarium head blight in wheat. Funct Integr Genomic 6:83–89. https://doi.org/10.1007/s10142-005-0007-y
Löffler M, Schön CC, Miedaner T (2009) Revealing the genetic architecture of FHB resistance in hexaploid wheat (Triticum aestivum L.) by QTL meta-analysis. Mol Breed 23:473–488. https://doi.org/10.1007/s11032-008-9250-y
Lorieux M (2012) MapDisto: fast and efficient computation of genetic linkage maps. Mol Breeding 30:1231–1235. https://doi.org/10.1007/s11032-012-9706-y
Maccaferri M, Cane MA, Sanguineti MC, Salvi S, Colalongo MC, Massi A, Clarke F, Knox R, Pozniak CJ, Clarke JM, Fahima T, Dubcovsky J, Xu S, Ammar K, Karsai I, Vida G, Tuberosa R (2014) A consensus framework map of durum wheat (Triticum durum Desf.) suitable for linkage disequilibrium analysis and genome-wide association mapping. BMC Genomics 15:873. https://doi.org/10.1186/1471-2164-15-873
McCaig T, DePauw R, Clarke J, McLeod J, Fernandez M, Knox R (1996) AC Barrie hard red spring wheat. Can J Plant Sci 76:337–339
McCartney CA, Brûlé-Babel AL, Fedak G, Martin RA, McCallum BD, Gilbert J, Hiebert CW, Pozniak CJ (2016) Fusarium head blight resistance QTL in the spring wheat cross Kenyon/86ISMN 2137. Front Microbiol 7:1542. https://doi.org/10.3389/fmicb.2016.01542
Mesterházy A (1995) Types and components of resistance to Fusarium head blight of wheat. Plant Breed 114:377–386. https://doi.org/10.1111/j.1439-0523.1995.tb00816.x
Miedaner T, Reinbrecht C, Lauber U, Schollenberger M, Geiger HH (2001) Effects of genotype and genotype-environment interaction on deoxynivalenol accumulation and resistance to Fusarium head blight in rye, triticale, and wheat. Plant Breeding 120:97–105. https://doi.org/10.1046/j.1439-0523.2001.00580.x
Miedaner T, Wilde F, Steiner B, Buerstmayr H, Korzun V, Ebmeyer E (2006) Stacking quantitative trait loci (QTL) for Fusarium head blight resistance from non-adapted sources in an European elite spring wheat background and assessing their effects on deoxynivalenol (DON) content and disease severity. Theor Appl Genet 112:562–569. https://doi.org/10.1007/s00122-005-0163-4
Oelke LM, Kolmer JA (2004) Characterization of leaf rust resistance in hard red spring wheat cultivars. Plant Dis 88:1127–1133. https://doi.org/10.1094/PDIS.2004.88.10.1127
Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, Sudhakar D, Christou P, Snape JW, Gale MD, Harberd NP (1999) 'Green revolution' genes encode mutant gibberellin response modulators. Nature 400:256–261. https://doi.org/10.1038/22307
Perez-Lara E, Semagn K, Chen H, Iqbal M, N'Diaye A, Kamran A, Navabi A, Pozniak C, Spaner D (2016) QTLs associated with agronomic traits in the Cutler × AC Barrie spring wheat mapping population using single nucleotide polymorphic markers. PLoS ONE. https://doi.org/10.1371/journal.pone.0160623
Rawat N, Pumphrey MO, Liu S, Zhang X, Tiwari VK, Ando K, Trick HN, Bockus WW, Akhunov E, Anderson JA, Gill BS (2016) Wheat Fhb1 encodes a chimeric lectin with agglutinin domains and a pore-forming toxin-like domain conferring resistance to Fusarium head blight. Nat Genet 48:1576–1580. https://doi.org/10.1038/ng.3706
Schroeder HW, Christensen JJ (1963) Factors affecting resistance of wheat to scab caused by Gibberella zeae. Phytopathology 53:831–838
Schweiger W, Steiner B, Ametz C, Siegwart G, Wiesenberger G, Berthiller F, Lemmens M, Jia H, Adam G, Muehlbauer GJ, Kreil DP, Buerstmayr H (2013) Transcriptomic characterization of two major Fusarium resistance quantitative trait loci (QTLs), Fhb1 and Qfhs.ifa-5A, identifies novel candidate genes. Mol Plant Pathol 14:772–785. https://doi.org/10.1111/mpp.12048
Schweiger W, Steiner B, Vautrin S, Nussbaumer T, Siegwart G, Zamini M, Jungreithmeier F, Gratl V, Lemmens M, Mayer KF, Berges H, Adam G, Buerstmayr H (2016) Suppressed recombination and unique candidate genes in the divergent haplotype encoding Fhb1, a major Fusarium head blight resistance locus in wheat. Theor Appl Genet 129:1607–1623. https://doi.org/10.1007/s00122-016-2727-x
Semagn K, Skinnes H, Bjørnstad Å, Marøy AG, Tarkegne Y (2007) Quantitative trait loci controlling fusarium head blight resistance and low deoxynivalenol content in hexaploid wheat population from ‘arina’ and NK93604. Crop Sci 47:294–303. https://doi.org/10.2135/cropsci2006.02.0095
Sherman JD, Yan L, Talbert L, Dubcovsky J (2004) A PCR marker for growth habit in common wheat based on allelic variation at the VRN-A1 gene. Crop Sci 44:1832–1838. https://doi.org/10.2135/cropsci2004.1832
Sinha RC, Savard ME, Lau R (1995) Production of monoclonal antibodies for the specific detection of deoxynivalenol and 15-acetyldeoxynivalenol by ELISA. J Agr Food Chem 43:1740–1744. https://doi.org/10.1021/jf00054a061
Somers DJ, Fedak G, Clarke J, Cao W (2006) Mapping of FHB resistance QTLs in tetraploid wheat. Genome 49:1586–1593. https://doi.org/10.1139/g06-127
Somers DJ, Fedak G, Savard M (2003) Molecular mapping of novel genes controlling Fusarium head blight resistance and deoxynivalenol accumulation in spring wheat. Genome 46:555–564. https://doi.org/10.1139/g03-033
Srinivasachary GN, Steed A, Hollins TW, Bayles R, Jennings P, Nicholson P (2009) Semi-dwarfing Rht-B1 and Rht-D1 loci of wheat differ significantly in their influence on resistance to Fusarium head blight. Theor Appl Genet 118:695–702. https://doi.org/10.1007/s00122-008-0930-0
Srinivasachary GN, Steed A, Simmonds J, Leverington-Waite M, Wang Y, Snape J, Nicholson P (2008) Susceptibility to Fusarium head blight is associated with the Rht-D1b semi-dwarfing allele in wheat. Theor Appl Genet 116:1145–1153. https://doi.org/10.1007/s00122-008-0742-2
Steiner B, Buerstmayr M, Wagner C, Danler A, Eshonkulov B, Ehn M, Buerstmayr H (2019) Fine-mapping of the Fusarium head blight resistance QTL Qfhs.ifa-5A identifies two resistance QTL associated with anther extrusion. Theor Appl Genet 132:2039–2053. https://doi.org/10.1007/s00122-019-03336-x
Steiner B, Lemmens M, Griesser M, Scholz U, Schondelmaier J, Buerstmayr H (2004) Molecular mapping of resistance to Fusarium head blight in the spring wheat cultivar Frontana. Theor Appl Genet 109:215–224. https://doi.org/10.1007/s00122-004-1620-1
Su Z, Bernardo A, Tian B, Chen H, Wang S, Ma H, Cai S, Liu D, Zhang D, Li T, Trick H, St Amand P, Yu J, Zhang Z, Bai G (2019) A deletion mutation in TaHRC confers Fhb1 resistance to Fusarium head blight in wheat. Nat Genet. https://doi.org/10.1038/s41588-019-0425-8
Su Z, Jin S, Zhang D, Bai G (2018) Development and validation of diagnostic markers for Fhb1 region, a major QTL for Fusarium head blight resistance in wheat. Theor Appl Genet. https://doi.org/10.1007/s00122-018-3159-6
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Wang S, Wong D, Forrest K, Allen A, Chao S, Huang BE, Maccaferri M, Salvi S, Milner SG, Cattivelli L, Mastrangelo AM, Whan A, Stephen S, Barker G, Wieseke R, Plieske J, Lillemo M, Mather D, Appels R, Dolferus R, Brown-Guedira G, Korol A, Akhunova AR, Feuillet C, Salse J, Morgante M, Pozniak C, Luo MC, Dvorak J, Morell M, Dubcovsky J, Ganal M, Tuberosa R, Lawley C, Mikoulitch I, Cavanagh C, Edwards KJ, Hayden M, Akhunov E, International Wheat Genome Sequencing Consortium (2014) Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol J 12:787–796. https://doi.org/10.1111/pbi.12183
Xiao J, Jin X, Jia X, Wang H, Cao A, Zhao W, Pei H, Xue Z, He L, Chen Q, Wang X (2013) Transcriptome-based discovery of pathways and genes related to resistance against Fusarium head blight in wheat landrace Wangshuibai. BMC Genomics 14:197. https://doi.org/10.1186/1471-2164-14-197
Xue S, Xu F, Tang M, Zhou Y, Li G, An X, Lin F, Xu H, Jia H, Zhang L, Kong Z, Ma Z (2011) Precise mapping Fhb5, a major QTL conditioning resistance to Fusarium infection in bread wheat (Triticum aestivum L.). Theor Appl Genet 123:1055–1063. https://doi.org/10.1007/s00122-011-1647-z
Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003) Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci U S A 100:6263–6268. https://doi.org/10.1073/pnas.0937399100
Yu J, Holland JB, McMullen MD, Buckler ES (2008) Genetic design and statistical power of nested association mapping in maize. Genetics 178:539–551. https://doi.org/10.1534/genetics.107.074245
Zhang W, Francis T, Gao P, Boyle K, Jiang F, Eudes F, Cuthbert R, Sharpe A, Fobert PR (2018) Genetic characterization of type II Fusarium head blight resistance derived from transgressive segregation in a cross between Eastern and Western Canadian spring wheat. Mol Breeding. https://doi.org/10.1007/s11032-017-0761-2
Acknowledgements
The authors thank technical staff from the participating laboratories for their contributions to this research. The authors wish to acknowledge Dr. Mark Jordan for providing laboratory facilities to carry out FHB candidate gene sequencing work. This project was funded by AAFC Growing Forward II, Western Grains Research Foundation, and as part of CTAG and CTAG2, Genome Prairie projects funded by Genome Canada, Manitoba Agriculture, Saskatchewan Ministry of Agriculture, and Western Grain Research Foundation.
Author information
Authors and Affiliations
Contributions
DT developed the linkage maps, conducted QTL analysis, characterized candidate genes, interpreted the data, and co-wrote the manuscript. AB, BB, GF, AF, JG, MH, DM, RM, BM, DS, MI, CP, and AN designed and conducted the field experiments and generated Fusarium head blight data. CM planned and organized the study, participated in the analysis and interpretation of data, and co-wrote the manuscript. All authors contributed to and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human and animal rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by Hermann Buerstmayr.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
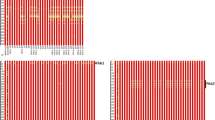
Supplementary file 2: Figure S1
CLUSTAL alignment of (a) DNA sequences and (b) deduced amino acid sequences of the predicted complete ORF of TaHRC gene amplified from a panel of wheat cultivars including three parents AC Barrie, Cutler, and Reeder using gene specific primer pair TaHRC-GSM-F/TaHRC-GSM-R (Su et al. 2018). Identical residues indicated by asterisks (*) and gaps are identified by dashes. Conserved amino acid substitutions are denoted with colon (:) and semi-conserved substitutions are indicated by a dot (.). Numbers on the right indicate the position number. The red rectangular frames show the locations of the start (ATG (+1)) and stop (TAA) codons, respectively. Fhb1-resistant haplotypes similar to Hap_Ning (Su et al. 2019) carried a large deletion including the start codon (ATG (+1)) and a 22 bp sequence downstream of the original ATG (shown in figure S1a), and part of upstream sequence (not shown in the Figure S1a) (PDF 134 kb)
Rights and permissions
About this article
Cite this article
Thambugala, D., Brûlé-Babel, A.L., Blackwell, B.A. et al. Genetic analyses of native Fusarium head blight resistance in two spring wheat populations identifies QTL near the B1, Ppd-D1, Rht-1, Vrn-1, Fhb1, Fhb2, and Fhb5 loci. Theor Appl Genet 133, 2775–2796 (2020). https://doi.org/10.1007/s00122-020-03631-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-020-03631-y