Abstract
Key message
We have mapped a new downy mildew resistance gene, Pl35, derived from wild Helianthus argophyllus to sunflower linkage group 1. New germplasms incorporating the Pl35 gene were developed for both oilseed and confection sunflower
Abstract
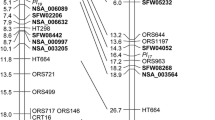
Sunflower downy mildew (DM), caused by the oomycete pathogen Plasmopara halstedii, is an economically important and widespread sunflower disease worldwide. Non-race-specific resistance is not available in sunflower, and breeding for DM resistance relies on race-specific resistance to control this disease. The discovery of the novel DM resistance genes is a long-term task due to the highly virulent and aggressive nature of the P. halstedii pathogen, which reduces the effectiveness of resistance genes. The objectives of this study were to: (1) transfer DM resistance from a wild sunflower species Helianthus argophyllus (PI 494576) into cultivated sunflowers; (2) map the resistance gene; and (3) develop diagnostic single-nucleotide polymorphism (SNP) markers for efficient targeting of the gene in breeding programs. The H. argophyllus accession PI 494576 previously identified with resistance to the most virulent P. halstedii race 777 was crossed with oilseed and confection sunflower in 2012. Molecular mapping using the BC2F2 and BC2F3 populations derived from the cross CONFSCLB1/PI 494576 located a new resistance gene Pl35 on linkage group 1 of the sunflower genome. The new gene Pl35 was successfully transferred from PI 494576 into cultivated sunflowers. SNP markers flanking Pl35 were surveyed in a validation panel of 548 diversified sunflower lines collected globally. Eleven SNP markers were found to be diagnostic for Pl35 SNP alleles, with four co-segregating with Pl35. The developed oilseed and confection germplasms with diagnostic SNP markers for Pl35 will be very useful resources for breeding of DM resistance in sunflower.




Similar content being viewed by others
References
Albourie JM, Tourvieille J, Tourvieille de Labrouhe D (1998) Resistance to metalaxyl in isolates of the sunflower pathogen Plasmopara halstedii. Eur J Plant Pathol 104:2335–2342
Bachlava E, Radwan OE, Abratti G, Tang S, Gao W, Heesacker AF, Bazzalo ME, Zambelli A, Leon AJ, Knapp SJ (2011) Downy mildew (Pl 8 and Pl 14) and rust (R Adv) resistance genes reside in close proximity to tandemly duplicated clusters of non-TIR-like NBS-LRR-encoding genes on sunflower chromosomes 1 and 13. Theor Appl Genet 122:1211–1221
Badouin H, Gouzy J, Grassa CJ, Murat F, Evan Staton S, Cottret L, Lelandais-Brière C, Owens GL, Carrère S, Mayjonade B, Legrand L, Gill N, Kane NC, Bowers JE, Hubner S, Bellec A, Bérard A, Bergès H, Blanchet N, Boniface MC, Brunel D, Catrice O, Chaidir N, Claudel C, Donnadieu C, Faraut T, Fievet G, Helmstetter N, King M, Knapp SJ, Lai Z, Le Paslier MC, Lippi Y, Lorenzon L, Mandel JR, Marage M, Marchand G, Marquand E, Bret-Mestries E, Morien E, Nambeesan S, Nguyen T, Pegot-Espagnet P, Pouilly N, Raftis F, Sallet E, Schiex T, Thomas J, Vandecasteele C, Varès D, Vear F, Vautrin S, Crespi M, Mangin B, Burke JM, Salse J, Muños S, Vincourt P, Rieseberg LH, Langlade NB (2017) The sunflower genome provides insights into oil metabolism, flowering and asterid evolution. Nature. https://doi.org/10.1038/nature22380
Bert PF, de Labrouhe TD, Philippon J, Mouzeyar S, Jouan I, Nicolas P, Vear F (2001) Identification of a second linkage group carrying genes controlling resistance to downy mildew (Plasmopara halstedii) in sunflower (Helianthus annuus L.). Theor Appl Genet 103:992–997
de Romano AB, Romano C, Bulos M, Altieri E, Sala C (2010) A new gene for resistance to downy mildew in sunflower. In: Proceedings of international symposium “sunflower breeding on resistance to diseases”, Krasnodar, Russia, June 23–24, 2010
Dußle CM, Hahn V, Knapp SJ, Bauer E (2004) Pl Arg from Helianthus argophyllus is unlinked to other known downy mildew resistance genes in sunflower. Theor Appl Genet 109:1083–1086
Ganal MW, Tanksley SD (1996) Recombination around the Tm2a and Mi resistance genes in different crosses of Lycopersicon peruvianum. Theor Appl Genet 92:101–108
Gascuel Q, Martinez Y, Boniface MC, Vear F, Pichon M, Godiard L (2015) The sunflower downy mildew pathogen Plasmopara halstedii. Mol Plant Pathol 16:109–122
Gedil MA, Slabaugh MB, Berry S, Segers B, Peleman J, Michelmore R, Miller JF, Gulya T, Knapp SJ (2001) Candidate disease resistance genes in sunflower cloned using conserved nucleotide binding site motifs: genetic mapping and linkage to downy mildew resistance gene Pl 1. Genome 44:205–212
Gilley M, Misar C, Gulya T, Markell S (2016) Prevalence and virulence of Plasmopara halstedii (downy mildew) in sunflowers. In: Proceeding 38th sunflower research forum. Fargo ND http://www.sunflowernsa.com/uploads/research/1277/Prevalence.Downey_Gilley.etal_2016.rev.pdf. Accessed 10 June 2019
Gulya TJ (1985) Registration of five disease—resistant sunflower germplasms. Crop Sci 25:719–720
Gulya TJ (2001) Field and greenhouse evaluations of new fungicides for the control of metalaxyl-resistant sunflower downy mildew. In: Proceeding 23rd sunflower research workshop. Fargo, ND. January 17–18, 2001, pp 29–34
Gulya TJ (2002) Efficacy of single and two-way fungicide seed treatments for the control of metalaxyl-resistant strains of Plasmopara halstedii (sunflower downy mildew). In: The BCPC conference—pests and diseases 2002. British Crop Protection Council. Proceedings brighton crop protection conference, November 18–21, 2002, Brighton, UK, pp. 575–580
Gulya TJ (2005) Evaluation of wild annual Helianthus species for resistance to downy mildew and Sclerotinia stalk rot. In: Proceeding 27th sunflower research forum. Fargo ND, 12–13 Jan 2005. http://www.sunflowernsa.com/uploads/research/265/GulyaWildHelianthus_studies_05.pdf
Gulya TJ, Carson ML, Urs RRNV (1982) Race 3 sunflower downy mildew: distribution and sources of resistance. Phytopathology 72:1136
Gulya TJ, Miller JF, Viranyi F, Sackston WE (1991a) Proposed internationally standardized methods for race identification of Plasmopara halstedii. Helia 14:11–20
Gulya TJ, Sackston WE, Viranyi F, Masirevic S, Rashid KY (1991b) New races of the sunflower downy mildew pathogen (Plasmopara halstedii) in Europe and North and South America. J Phytopathol 132:303–311
Gulya TJ, Draper M, Harbour J, Holen C, Knodel J, Lamey A, Mason P (1999) Metalaxyl resistance in sunflower downy mildew in North America. In: Proceedings of 21st sunflower research workshop. Fargo ND, January 14–15, 1999, pp 118–123
Gulya TJ, Markell S, McMullen M, Harveson B, Osborne L (2011) New virulent races of downy mildew: distribution, status of DM resistant hybrids, and USDA sources of resistance. In: Proceedings of 33th sunflower research forum. Fargo ND, Jan 12–13, 2011. http://www.sunflowernsa.com/uploads/17/gulya_virulentracesdownymildew.pdf
Hladni N (2016) Present status and future prospects of global confectionary sunflower production. In: Proceedings of 19th international sunflower conference. Edirne, Turkey, May 29–June 3, 2016, pp 47–60
Hulke BS, Miller JF, Gulya TJ, Vick BA (2010) Registration of the oilseed sunflower genetic stocks HA 458, HA 459, and HA 460 possessing genes for resistance to downy mildew. J Plant Regist 4:93–97
Hulke BS, Ma G, Qi LL, Gulya TJ (2018) Registration of oilseed sunflower germplasms RHA 461, RHA 462, RHA 463, HA 465, HA 466, HA 467, RHA 468 with diversity in Sclerotinia resistance, yield, and other traits. J Plant Regist 12:142–147
Jan CC (1992) Four sunflower nuclear male-sterile genetic stocks. Crop Sci 32:1519
Jan CC, Gulya TJ (2006) Registration of a sunflower germplasm resistant to rust, downy mildew and virus. Crop Sci 46:1829
Järve K, Peusha HO, Tsymbalova J, Tamm S, Devos KM, Enno TM (2000) Chromosome location of a Tritivum timopheevii-derived powdery mildew resistance gene transferred to common wheat. Genome 46:70–82
Liu Z, Gulya TJ, Seiler GJ, Vick BA, Jan CC (2012) Molecular mapping of the Pl 16 downy mildew resistance gene from HA-R4 to facilitate marker-assisted selection in sunflower. Theor Appl Genet 125:121–131
Liu Z, Zhang L, Ma GJ, Seiler GJ, Jan CC, Qi LL (2019) Molecular mapping of the downy mildew and rust resistance genes in a sunflower germplasm line TX16R. Mol Breed 39:19
Livaja M, Wang Y, Wieckhorst S, Haseneyer G, Hahn V, Knapp SJ, Taudien S, Schön C-C, Bauer E (2013) BSTA: a targeted approach combines bulked segregant analysis with Next-generation sequencing and de novo transcriptome assembly for SNP discovery in sunflower. BMC Genom 14:628
Long YM, Chao WS, Ma GJ, Xu SS, Qi LL (2017) An innovative SNP genotyping method adapting multiple platforms and throughputs. Theor Appl Genet 130:597–607
Ma GJ, Markell SG, Song QJ, Qi LL (2017) Genotyping-by-sequencing targeting of a novel downy mildew resistance gene Pl 20 from wild Helianthus argophyllus in sunflower (Helianthus annuus L.). Theor Appl Genet 130:1519–1529
Ma GJ, Seiler GJ, Markell SG, Qi LL (2019) Registration of three confection sunflower germplasms, HA-DM2, HA-DM3 and HA-DM4, resistant to downy mildew and rust. Plant Regist 13:103–108
Markell SG, Humann RH, Gilley M, Gulya TJ (2016) Downy mildew pathogen. In: Harveson RM, Markell SG, Block CC, Gulya TJ (eds) Compendium of sunflower diseases and pests. American Phytopathology Press, St. Paul, pp 15–117
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Miller JF, Gulya TJ (1987) Inheritance of resistance to race 3 downy mildew in sunflower. Crop Sci 27:210–212
Miller JF, Gulya TJ (1991) Inheritance of resistance to race 4 of downy mildew derived from interspecific crosses in sunflower. Crop Sci 31:40–43
Miller JF, Gulya TJ, Seiler GJ (2002) Registration of five fertility restorer sunflower germplasms. Crop Sci 42:989–991
Miller JF, Gulya TJ, Vick BA (2006) Registration of three maintainer (HA 444 to HA 446) and one restorer (RHA 467) high oleic oilseed sunflower germplasms. Crop Sci 46:484–485
Molinero-Ruiz ML, Melero-Vara JM, Dominguez J (2003) Inheritance of resistance to two races of sunflower downy mildew (Plasmopara halstedii) in two Helianthus annuus L. lines. Euphytica 131:47–51
Mulpuri S, Liu Z, Feng J, Gulya TJ, Jan CC (2009) Inheritance and molecular mapping of a downy mildew resistance gene, Pl 13 in cultivated sunflower (Helianthus annuus L.). Theor Appl Genet 119:795–803
Neu C, Stein N, Keller B (2002) Genetic mapping of the Lr20-Pm1 resistance locus reveals suppressed recombination on chromosome arm 7AL in hexaploid wheat. Genome 45:737–744
Nishimura M (1922) Study in Plasmopara halstedii. J Coll Agric Hokkaido Imp Univ XI(Pt 3):185–210
Pecrix Y, Buendia L, Penouilh-Suzette C, Maréchaux M, Legrand L, Bouchez Q, Rengel D, Gouzy J, Cottret L, Vear F, Godiard L (2018a) Sunflower resistance to multiple downy mildew pathotypes revealed by recognition of conserved effectors of the oomycete Plasmopara halstedii. Plant J. https://doi.org/10.1111/tpj.14157
Pecrix Y, Penouilh-Suzette C, Stéphane Muños S, Felicity Vear F, Godiard L (2018b) Ten broad spectrum resistances to downy mildew physically mapped on the sunflower genome. Front Plant Sci 9:1780. https://doi.org/10.3389/fpls.2018.01780
Pegadaraju V, Nipper R, Hulke BS, Qi LL, Schultz Q (2013) De novo sequencing of the sunflower genome for SNP discovery using the RAD (Restriction site Associated DNA) approach. BMC Genom 14:556
Qi LL, Gulya TJ, Seiler GJ, Hulke BS, Vick BA (2011) Identification of resistance to new virulent races of rust in sunflowers and validation of DNA markers in the gene pool. Phytopathology 101:241–249
Qi LL, Long YM, Jan CC, Ma GJ, Gulya TJ (2015) Pl 17 is a novel gene independent of known downy mildew resistance genes in the cultivated sunflower (Helianthus annuus L.). Theor Appl Genet 128:757–767
Qi LL, Foley ME, Cai XW, Gulya TJ (2016) Genetics and mapping of a novel downy mildew resistance gene, Pl 18, introgressed from wild Helianthus argophyllus into cultivated sunflower (Helianthus annuus L.). Theor Appl Genet 129:741–752
Qi LL, Talukder ZI, Hulke BS, Foley ME (2017) Development and dissection of diagnostic SNP markers for the downy mildew resistance genes Pl Arg and Pl 8 and maker-assisted gene pyramiding in sunflower (Helianthus annuus L.). Mol Genet Genom 292:551–563
Radwan O, Bouzidi MF, Vear F, Philippon J, de Labrouhe TD, Nicolas P, Mouzeyar S (2003) Identification of non-TIR-NBS-LRR markers linked to the Pl 5/Pl 8 locus for resistance to downy mildew in sunflower. Theor Appl Genet 106:1438–1446
Rahim M, Jan CC, Gulya TJ (2002) Inheritance of resistance to sunflower downy mildew races 1, 2 and 3 in cultivated sunflower. Plant Breed 121:57–60
Roeckel-Drevet P, Gagne G, Mouzeyar S, Gentzbittel L, Philippon J, Nicolas P, de Labrouhe DT, Vear F (1996) Colocation of downy mildew (Plasmopara halstedii) resistance genes in sunflower (Helianthus annuus L.). Euphytica 91:225–228
Seiler GJ (1991) Registration of 13 downy mildew tolerant interspecific sunflower germplasm lines derived from wild annual species. Crop Sci 31:1714–1716
Seiler GJ, Qi LL, Marek LF (2017) Utilization of sunflower crop wild relatives for cultivated sunflower improvement. Crop Sci. https://doi.org/10.2135/cropsci2016.10.0856
Slabaugh MB, Yu JK, Tang SX, Heesacker A, Hu X, Lu GH, Bidney D, Han F, Knapp SJ (2003) Haplotyping and mapping a large cluster of downy mildew resistance gene candidates in sunflower using multilocus intron fragment length polymorphisms. Plant Biotechnol J 1:167–185
Stirling B, Newcombe G, Vrebalov J, Bosdet I, Bradshaw HD Jr (2001) Suppressed recombination around the MXC3 locus, a major gene for resistance to poplar leaf rust. Theor Appl Genet 103:1129–1137
Talukder ZI, Gong L, Hulke BS, Pegadaraju V, Song QJ, Schultz Q, Qi LL (2014) A high-density SNP map of sunflower derived from RAD-sequencing facilitating fine-mapping of the rust resistance gene R 12. PLoS One 9:e98628
Talukder ZI, Ma GJ, Hulke BS, Jan CC, Qi LL (2019) Linkage mapping and genome-wide association studies of the Rf gene cluster in sunflower (Helianthus annuus L.) and their distribution in world sunflower collections. Front Genet 10:216
Tang S, Yu JK, Slabaugh MB, Shintani DK, Knapp SJ (2002) Simple sequence repeat map of the sunflower genome. Theor Appl Genet 105:1124–1136
Tang S, Kishore VK, Knapp SJ (2003) PCR-multiplexes for a genome-wide framework of simple sequence repeat marker loci in cultivated sunflower. Theor Appl Genet 107:6–19
Trojanova Z, Sedlarova M, Gulya TJ, Lebeda A (2016) Methodology of virulence screening and race characterization of Plasmopara halstedii, and resistance evaluation in sunflower—a review. Plant Pathol. https://doi.org/10.1111/ppa.12593
Van Daelen R, Gerbens F, Van Ruissen F, Arts J, Hontelez J, Zabel P (1993) Long-range physical maps of two loci (Aps-1 and GP79) flanking the root-knot nematode resistance gene (Mi) near the centromere of tomato chromosome 6. Plant Mol Biol 23:186–192
Van Ooijen JW (2006) JoinMap ® 4, Software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen, Netherlands. http://www.kyazma.com. Accessed 10 June 2019
Vear F (1974) Studies on resistance to downy mildew in sunflower (Helianthus annuus L.). In: Proceedings of 6th international sunflower conference, 22–24 July, 1974, Bucharest, Romania, pp 297–302
Vear F (2016) Changes in sunflower breeding over the last fifty years. OCL 23:D202
Vear F, Leclercq P (1971) Deux nouvea ux genes resistance au mildiou du tournesol. Ann Amelior Plant 21:251–255
Vear F, Gentzbittel L, Philippon J, Mouzeyar S, Mestries E, Roeckel-Drevet P, de Labroube DT, Nicolas P (1997) The genetics of resistance to five races of downy mildew (Plasmopara halstedii) in sunflower (Helianthus annuus L.). Theor Appl Genet 95:584–589
Vear F, Serre F, Jouan-Dufournel I, Bert PF, Roche S, Walser P, Tourvieille de Labrouhe D, Vincourt P (2008a) Inheritance of quantitative resistance to downy mildew (Plasmopara halstedii) in sunflower (Helianthus annuus L.). Euphytica 164:561–570
Vear F, Seriveys H, Petit A, Serre F, Boudon JP, Roche S, Walser P, de Labrouhe DT (2008b) Origins of major genes for downy mildew resistance in sunflower. In: Proceedings of 17th international sunflower conference Cordoba, Spain, 2008, pp 125–130
Vincourt P, As-sadi F, Bordat A, Langlade NB, Gouzy J, Pouilly N, Lippi Y, Serre F, Godiard L, Tourvieille de Labrouhe D, Vear F (2012) Consensus mapping of major resistance genes and independent QTL for quantitative resistance to sunflower downy mildew. Theor Appl Genet 125:909–920
Viranyi F, Gulya TJ, Tourieille DL (2015) Recent changes in the pathogenic variability of Plasmopara halstedii (sunflower downy mildew) populations from different continents. Helia 38:149–162
Vrânceanu V, Stoenescu F (1970) Immunity to sunflower downy mildew due to a single dominant gene. Probl Agric 22:34–40
Wei F, Gobelman-Werner K, Morroll SM, Kurth J, Mao L, Wing R, Leister D, Schulze-Lefert P, Wise RP (1999) The Mla (Powdery Mildew) resistance cluster is associated with three NBS-LRR gene families and suppressed recombination within a 240-kb DNA interval on chromosome 5S (1HS) of barley. Genetics 153:1929–1948
Wieckhorst S, Bachlava E, Dußle CM, Tang S, Gao W, Saski C, Knapp SJ, Schön CC, Hahn V, Bauer E (2010) Fine mapping of the sunflower resistance locus Pl ARG introduced from the wild species Helianthus argophyllus. Theor Appl Genet 121:1633–1644
Xie W, Ben-David R, Zeng B, Dinoor A, Xie C, Sun Q, Röder MS, Fahoum A, Fahima T (2012) Suppressed recombination rate in 6VS/6AL translocation region carrying the Pm21 locus introgressed from Haynaldia villosa into hexaploid wheat. Mol Breed 29:399–412
Young PA, Morris HE (1927) Plasmopara downy mildew of cultivated sunflower. Am J Bot 14:551–558
Yu JK, Tang S, Slabaugh MB, Heesacker A, Cole G, Herring M et al (2003) Towards a saturated molecular genetic linkage map for cultivated sunflower. Crop Sci 43:367–387
Zhang ZW, Ma GJ, Zhao J, Markell SG, Qi LL (2017) Discovery and introgression of the wild sunflower-derived novel downy mildew resistance gene Pl 19 in confection sunflower (Helianthus annuus L.). Theor Appl Genet 130:29–39
Zimmer DE (1974) Physiological specialization between races of Plasmopara halstedii in America and Europe. Phytopathology 64:1465–1467
Zimmer DE, Kinman ML (1972) Downy mildew resistance in cultivated sunflower and its inheritance. Crop Sci 12:749–751
Acknowledgements
We thank Angelia Hogness for technical assistance in the laboratory, greenhouse, and field. This project was supported by the USDA-ARS CRIS Project No. 5442-21000-043-00D and the USDA-AMS Specialty Crop Block Grant Program 12-25-B-1689. Mention of trade names or commercial products in this report is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. The USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Contributions
LLQ conceived and designed the experiments, analyzed the data, and wrote the paper. LLQ, GJM, XHL, and GJS performed the experiments. GJS, GJM, and XHL commented on the manuscript before submission.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The experiments were performed in compliance with the current laws of the USA.
Additional information
Communicated by Beat Keller.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Qi, L.L., Ma, G.J., Li, X.H. et al. Diversification of the downy mildew resistance gene pool by introgression of a new gene, Pl35, from wild Helianthus argophyllus into oilseed and confection sunflowers (Helianthus annuus L.). Theor Appl Genet 132, 2553–2565 (2019). https://doi.org/10.1007/s00122-019-03370-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-019-03370-9