Abstract.
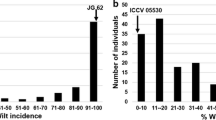
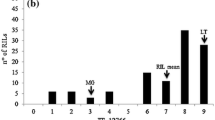
Ascochyta blight is an economically important disease of chickpea caused by the fungus Ascochyta rabiei. The fungus shows considerable variation for pathogenicity in nature. However, studies on the genetics of pathotype-specific resistance are not available for this plant-pathosystem. The chickpea landrace ILC 3279 has resistance to pathotype I and II of the pathogen. In order to understand the inheritance of pathotype-specific resistance in this crop, both Mendelian and quantitative trait loci analyses were performed using a set of intraspecific, recombinant inbred lines derived from a cross between the susceptible accession ILC 1272 and the resistant ILC 3279, and microsatellite markers. We identified and mapped a major locus (ar1, mapped on linkage group 2), which confers resistance to pathotype I, and two independent recessive major loci (ar2a, mapped on linkage group 2 and ar2b, mapped on linkage group 4), with complementary gene action conferring resistance to pathotype II. Out of two pathotype II-specific resistance loci, one (ar2a) linked very closely with the pathotype I-specific resistance locus, indicating a clustering of resistance genes in that region of the chickpea genome.



Similar content being viewed by others
References
Anderson PA, Okubura PA, Arroyogarcia R, Meyers BC, Michelmore RW (1996) Molecular analysis of irradiation-induced and spontaneous deletion mutants at a disease resistance locus in Lactuca sativa. Mol Gen Genet 251:316–325
Banerjee D, Zhang X, Bent AF (2001) The leucine-rich repeat domain can determine effective interaction between RPS2 and other host factors in Arabidopsis RPS2-mediated disease resistance. Genetics 158:439–450
Dickinson MJ, Jones DA, Jones JDG (1993) Close linkage between the Cf-2/Cf-5 and Mi resistance loci in tomato. Mol Plant-Microbe Interact 6:341–347
Dixon MS, Jones DA, Keddie JS, Thomas CM, Harrison K, Jones JD (1996) The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84:451–459
Dixon MS, Hatzixanthis K, Jones DA, Harrison K, Jones JDG (1998) The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell 10:1915–1925
Ghislain M, Trognitz B, Herrera MR, Solis J, Casallo G, Vásquez C, Hurtado O, Castillo R, Portal L, Orrillo M (2001) Genetic loci associated with field resistance to late blight in offspring of Solanum phureja and S. tuberosum grown under short-day conditions. Theor Appl Genet 103:33–442
Hüttel B, Winter P, Weising K, Choumane W, Weigand F, Kahl G (1999) Sequence-tagged microsatellite-site markers for chickpea (Cicer arietinum L.). Genome 42:210–217
Iqbal MJ, Meksem K, Njiti VN, Kassem MA, Lightfoot DA (2001) Microsatellite markers identify three additional quantitative trait loci for resistance to soybean sudden-death syndrome (SDS) in Essex × Forrest RILs. Theor Appl Genet 102:187–192
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Kusmenoglu I (1990) Ascochyta blight of chickpea: inheritance and relationship to seed size, morphological traits and isozyme variation. MS thesis, Washington State University, Pullman, Washington
Lincoln SE, Daly MJ, Lander ES (1993) MAPMAKER/EXP version 3.0: A Tutorial and Reference Manual, 3rd edn. Whitehead Institute for Biomedical Research, Cambridge, Massachusetts, USA
Mackill DJ, Bonman JM (1992) Inheritance of blast resistance in near-isogenic lines of rice. Phytopathology 82:746–749
Maddala GS (1992) Introduction to Econometrics, 2nd edn. MacMillan Publishing Company, New York, pp 119–120
Malik BA, Rahman MS (1992) Breeding for ascochyta blight-resistance desi chickpea in Pakistan. In: Singh KB, Saxena MC (eds) Disease resistance breeding in chickpea. ICARDA, Aleppo, Syria, pp 77–85
Meyers BC, Chin DB, Shen KA, Sivaramakrishnan S, Lavelle DO, Zhang Z, Michelmore RW (1998) The major resistance gene cluster in lettuce is highly duplicated and spans several megabases. Plant Cell 10:1817–1832
Nene YL, Reddy MV (1987) Chickpea diseases and their control. In: Singh KB, Saxena MC (eds) Disease resistance breeding in chickpea. ICARDA, Aleppo, Syria, pp 233–270
Parniske M, Hammond-Kosack KE, Golstein C, Thomas CM, Jones DA, Harrison K, Wulff BB, Jones JD (1997) Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91:821–832
Porta-Puglia A (1992) Variability in Ascochyta rabiei. In: Singh KB, Saxena MC (eds) Disease resistance breeding in chickpea. ICARDA, Aleppo, Syria, pp 135–143
Reddy MV, Nene YL, Singh G, Bashir M (1990) Strategies for management of foliar diseases of chickpea. In:Chickpea in the nineties. Proc 2nd Int Workshop on Chickpea Improvement, 4–8 Dec. 1989, ICRISAT Center, India, pp 117–127
Santra DK, Tekeoglu M, Ratnaparkhe M, Kaiser WJ, Muehlbauer FJ (2000) Identification and mapping of QTLs conferring resistance to ascochyta blight in chickpea. Crop Sci 40:1606–1612
Shen KA, Meyers BC, Islam-Faridi MN, Chin DB, Stelly DM, Michelmore RW (1998) Resistance gene candidates identified by PCR with degenerate oligonucleotide primers map to clusters of resistance genes in lettuce. Mol Plant-Microbe Interact 11:815–823
Singh KB, Reddy MV (1983) Inheritance of resistance to ascochyta blight in chickpea. Crop Sci 23:9–10
Singh KB, Reddy MV (1993) Resistance to six races of Ascochyta rabiei in the world germplasm collection of chickpea. Crop Sci 33:186–189
Singh KB, Hawtin GC, Nene YL, Reddy MV (1981) Resistance in chickpeas to Ascochyta rabiei. Plant Dis 65:586–587
Singh KB, Reddy MV, Haware MP (1992) Breeding for resistance to ascochyta blight in chickpea. In: Singh KB, Saxena MC (eds) Disease resistance breeding in chickpea. ICARDA, Aleppo, Syria, pp 23–54
Song WY, Pi LY, Wang GL, Gardner J, HolstenT, Ronald PC (1997) Evolution of the rice Xa21 disease resistance gene family. Plant Cell 9:1279–1287
Tekeoglu M, Santra DK, Kaiser WJ, Muehlbauer FJ (2000) Ascochyta blight resistance in three chickpea recombinant inbred line populations. Crop Sci 40:1251–1256
Tewari SK, Pandey MP (1986) Genetics of resistance to ascochyta blight of chickpea (Cicer arietinum L.). Euphytica 35:211–215
Udupa SM, Weigand F, Saxena MC, Kahl G (1998) Genotyping with RAPD and microsatellite markers resolves pathotype diversity in the ascochyta blight pathogen of chickpea. Theor Appl Genet 97:299–307
Udupa SM, Robertson LD, Weigand F, Baum M, Kahl G (1999) Allelic variation at (TAA) n microsatellite loci in a world collection of chickpea (Cicer arietinum L.) germplasm. Mol Gen Genet 261:354–363
Vir S, Grewal JS (1974) Physiological specialization in Ascochyta rabiei, the causal organism of gram blight. Indian Phytopathol 27:265–266
Vir S, Grewal JS, Gupta VP (1975) Inheritance of resistance to ascochyta blight in chickpea. Euphytica 24:209–211
Wang D, Arelli PR, Shoemaker RC, Diers BW (2001) Loci underlying resistance to Race 3 of soybean cyst nematode in Glycine soja plant introduction 468916. Theor Appl Genet 103:561–566
Wang GL, Mackill DJ, Bonman JM, McCouch SR, Champoux MC, Nelson RJ (1994) RFLP mapping of genes conferring complete and partial resistance to blast from durably resistant rice cultivar. Genetics 136:1421–1434
Wilson IW, Schiff CL, Hughes DE, Somerville SC (2001) Quantitative trait loci analysis of powdery mildew disease resistance in the Arabidopsis thaliana accession Kashmir-1. Genetics 158:1301–1309
Winter P, Pfaff T, Udupa SM, Hüttel B, Sharma PC, Sahi S, Arreguin-Espinoza R, Weigand F, Muehlbauer FJ, Kahl G (1999) Characterization and mapping of sequence-tagged microsatellite sites in the chickpea (Cicer arietinum L.) genome. Mol Gen Genet 262:90–101
Winter P, Beko-Iseppon A.-M, Hüttel B, Ratnaparkhe M, Tullu A, Sonnante G, Pfaff T, Tekeoglu M, Santra D, Sant VJ, Rajesh PN, Kahl G, Muehlbauer FJ (2000) A linkage map of the chickpea (Cicer arietinum L.) genome based on recombinant inbred lines from a C. arietinum × C. reticulatum cross: localization of resistance genes for Fusarium wilt races 4 and 5. Theor Appl Genet 101:1155–1163
Wright RJ, Thaxton PM, El-Zik, KM, Paterson AH (1998) D-subgenome bias of Xcm resistance genes in tetraploid gossypium (cotton) suggests that polyploid formation has created novel avenues for evolution. Genetics 149:1987–1996
Young ND (1996) QTL mapping and quantitative disease resistance in plants. Annu Rev Phytopathol 34:479–501
Acknowledgements.
The authors' research was supported by grants to ICARDA from the German Federal Ministry of Economic Cooperation and Development (BMZ, Bonn, Germany) and the Arab Fund for Economic and Social Development (AFESD, Kuwait).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Hagemann
Rights and permissions
About this article
Cite this article
Udupa, S.M., Baum, M. Genetic dissection of pathotype-specific resistance to ascochyta blight disease in chickpea (Cicer arietinum L.) using microsatellite markers. Theor Appl Genet 106, 1196–1202 (2003). https://doi.org/10.1007/s00122-002-1168-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-002-1168-x