Abstract
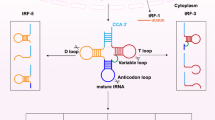
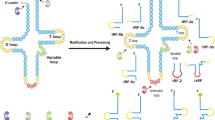
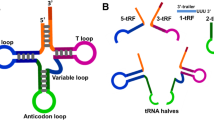
The number of studies on non-coding RNAs has increased substantially in recent years owing to their importance in gene regulation. However, the biological functions of small RNAs from abundant species of housekeeping non-coding RNAs (rRNA, tRNA, etc.) remain a highly studied topic. tRNA-derived small RNAs (tsRNAs) refer to the specific cleavage of tRNAs by specific nucleases [e.g., Dicer and angiogenin (ANG)] in particular cells or tissues or under certain conditions such as stress and hypoxia. tsRNAs are a type of non-coding small RNA that are widely found in the prokaryotic and eukaryotic transcriptomes and are generated from mature tRNAs or precursor tRNAs at different sites. There are two main types of tsRNAs, tRNA-derived fragments (tRFs) and tRNA halves. tRFs are 14–30 nucleotides (nt) long and mainly consist of three subclasses: tRF-5, tRF-3, and tRF-1. tRNA halves, which are 31–40 nt long, are generated by specific cleavage in the anticodon loops of mature tRNAs. There are two types of tRNA halves, 5′-tRNA halves and 3′-tRNA halves. tsRNAs have multiple biological functions including acting as signaling molecules in stress responses and as regulators of gene expression. Additionally, they have been considered to be involved in RNA processing, cell proliferation, translation suppression, the modulation of DNA damage response, and neurodegeneration. More importantly, they are closely related to the occurrence of many human diseases such as tumors, infectious diseases, metabolic diseases, and neurological diseases. Moreover, tsRNAs have the potential to become new biomarkers for disease diagnosis. Continuous investigations will help us to understand their generation and regulatory mechanisms as well as the possible roles of tRFs and tRNA halves.

Similar content being viewed by others
References
Sun W, Yang Y, Xu C, Guo J (2017) Regulatory mechanisms of long noncoding RNAs on gene expression in cancers. Cancer Genet 216-217:105–110
Shaker OG, Mohammed SR, Mohammed AM, Mahmoud Z (2018) Impact of microRNA-375 and its target gene SMAD-7 polymorphism on susceptibility of colorectal cancer. J Clin Lab Anal 32(1):e22215
Cole C, Sobala A, Cheng L, Thatcher SR, Bowman A, Brown JWS, Green PJ, Barton GJ, Hutvagner G (2009) Filtering of deep sequencing data reveals the existence of abundant dicer-dependent small RNAs derived from tRNAs. RNA 15(12):2147–2160
Tian M, Chen R, Li T, Xiao B (2018) Reduced expression of circRNA hsa_circ_0003159 in gastric cancer and its clinical significance. J Clin Lab Anal 32(3):e22281
Li T, Shao Y, Fu L, Xie Y, Zhu L, Sun W, Yu R, Xiao B, Guo J (2018) Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med (Berl) 96(1):85–96
Nie ZL, Wang YS, Mei YP, Lin X, Zhang GX, Sun HL, Wang YL, Xia YX, Wang SK (2018) Prognostic significance of long noncoding RNA Z38 as a candidate biomarker in breast cancer. J Clin Lab Anal 32(1):e22193
Zhu LW, Xie Y, Guo JM (2017) The biological functions of tRNA-derived fragments and tRNA halves, and their roles in the pathogenesis. Prog Biochem Biophys 44(7):565–572
Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, Dalla-Favera R (2013) tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A 110(4):1404–1409
Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, Zheng X (2009) Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett 583(2):437–442
Li S, Xu Z, Sheng J (2018) tRNA-derived small RNA: a novel regulatory small non-coding RNA. Genes (Basel) 9(5):e246
Kuscu C, Kumar P, Kiran M, Su Z, Malik A, Dutta A (2018) tRNA fragments (tRFs) guide ago to regulate gene expression post-transcriptionally in a dicer independent manner. RNA 24:1093–1105
Shigematsu M, Kirino Y (2015) tRNA-derived short non-coding RNA as interacting partners of Argonaute proteins. Gene Regul Syst Biol 109:27–33
Olvedy M, Scaravilli M, Hoogstrate Y, Visakorpi T, Jenster G, Martens-Uzunova E (2016) A comprehensive repertoire of tRNA-derived fragments in prostate cancer. Oncotarget 7(17):24766–24777
Kumar P, Mudunuri SB, Anaya J, Dutta A (2015) tRFdb: a database for transfer RNA fragments. Nucleic Acids Res 43:D141–D145
Gupta N, Singh A, Zahra S, Kumar S (2018) PtRFdb: a database for plant transfer RNA-derived fragments. Database (Oxford) 2018: bay063
Loher P, Telonis AG, Rigoutsos I (2018) Accurate profiling and quantification of tRNA fragments from RNA-seq data: a vade mecum for MINTmap. Methods Mol Biol 1680:237–255
Grelet S, McShane A, Hok E, Tomberlin J, Howe PH, Geslain R (2017) SPOt: a novel and streamlined microarray platform for observing cellular tRNA levels. PLoS One 12(5):e0177939
Couvillion MT, Sachidanandam R, Collins K (2010) A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes Dev 24:2742–2747
Veneziano D, Di Bella S, Nigita G, Laganà A, Ferro A, Croce CM (2016) Noncoding RNA: current deep sequencing data analysis approaches and challenges. Hum Mutat 37(12):1283–1298
Lee YS, Shibata Y, Malhotra A, Dutta A (2009) A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 23:2639–2649
Kumar P, Anaya J, Mudunuri SB, Dutta A (2014) Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol 12:78
Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF (2015) Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell 161:790–802
Schaffer AE, Eggens VR, Caglayan AO, Reuter MS, Scott E, Coufal NG, Silhavy JL, Xue Y, Kayserili H, Yasuno K et al (2014) CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. Cell 157:651–663
Kumar P, Kuscu C, Dutta A (2016) Biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem Sci 41(8):679–689
Hanada T, Weitzer S, Mair B, Bernreuther C, Wainger BJ, Ichida J, Hanada R, Orthofer M, Cronin SJ, Komnenovic V, Minis A, Sato F, Mimata H, Yoshimura A, Tamir I, Rainer J, Kofler R, Yaron A, Eggan KC, Woolf CJ, Glatzel M, Herbst R, Martinez J, Penninger JM (2013) CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature 495:474–480
Karaca E, Weitzer S, Pehlivan D, Shiraishi H, Gogakos T, Hanada T, Jhangiani SN, Wiszniewski W, Withers M, Campbell IM, Erdin S, Isikay S, Franco LM, Gonzaga-Jauregui C, Gambin T, Gelowani V, Hunter JV, Yesil G, Koparir E, Yilmaz S, Brown M, Briskin D, Hafner M, Morozov P, Farazi TA, Bernreuther C, Glatzel M, Trattnig S, Friske J, Kronnerwetter C, Bainbridge MN, Gezdirici A, Seven M, Muzny DM, Boerwinkle E, Ozen M, Baylor Hopkins Center for Mendelian Genomics, Clausen T, Tuschl T, Yuksel A, Hess A, Gibbs RA, Martinez J, Penninger JM, Lupski JR (2014) Human CLP1 mutations alter tRNA biogenesis, affecting both peripheral and central nervous system function. Cell 157:636–650
Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA (2010) Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16(4):673–695
Yamasaki S, Ivanov P, Hu GF, Anderson P (2009) Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 185(1):35–42
Li S, Hu GF (2012) Emerging role of angiogenin in stress response and cell survival under adverse conditions. J Cell Physiol 227:2822–2826
Saikia M, Krokowski D, Guan BJ, Ivanov P, Parisien M, Hu GF, Anderson P, Pan T, Hatzoglou M (2012) Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J Biol Chem 287:42708–42725
Pagès A, Dotu I, Pallarès-Albanell J, Martí E, Guigó R, Eyras E (2018) The discovery potential of RNA processing profiles. Nucleic Acids Res 46(3):e15
Pundhir S, Gorodkin J (2015) Differential and coherent processing patterns from small RNAs. Sci Rep 5:12062
Yeung ML, Bennasser Y, Watashi K, Le SY, Houzet L, Jeang KT (2009) Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res 37:6575–6586
Reese TA, Xia J, Johnson LS, Zhou X, Zhang W, Virgin HW (2010) Identification of novel microRNA-like molecules generated from herpesvirus and host tRNA transcripts. J Virol 84:10344–10353
Heyer R, Dörr M, Jellen-Ritter A, Späth B, Babski J, Jaschinski K, Soppa J, Marchfelder A (2012) High throughput sequencing reveals a plethora of small RNAs including tRNA derived fragments in Haloferax volcanii. RNA Biol 9:1011–1018
Couvillion MT, Bounova G, Purdom E, Speed TP, Collins K (2012) A Tetrahymena Piwi bound to mature tRNA 30 fragments activates the exonuclease Xrn2 for RNA processing in the nucleus. Mol Cell 48:509–520
Chen CJ, Liu Q, Zhang YC, Qu LH, Chen YQ, Gautheret D (2011) Genome-wide discovery and analysis of microRNAs and other small RNAs from rice embryogenic callus. RNA Biol 8:538–547
Hackenberg M, Huang PJ, Huang CY, Shi BJ, Gustafson P, Langridge P (2012) A comprehensive expression profile of microRNAs and other classes of non-coding small RNAs in barley under phosphorous-deficient and -sufficient conditions. DNA Res 20:109–125
Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R (2008) Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, dicer-dependent small RNAs. Genes Dev 22:2773–2785
Li Z, Ender C, Meister G, Moore PS, Chang Y, John B (2012) Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res 40:6787–6799
Pederson T (2010) Regulatory RNAs derived from transfer RNA? RNA 16:1865–1869
Karaiskos S, Naqvi AS, Swanson KE, Grigoriev A (2015) Age-driven modulation of tRNA-derived fragments in Drosophila and their potential targets. Biol Direct 16(10):51
Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, Daub CO, Kai C, Kawai J, Yasuda J et al (2008) Hidden layers of human small RNAs. BMC Genomics 9:157
Bidartondo MI (2008) Preserving accuracy in GenBank. Science 319(5870):1616
Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P (2011) Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell 43(4):613–623
Sobala A, Hutvagner G (2013) Small RNAs derived from the 50 end of tRNA can inhibit protein translation in human cells. RNA Biol 10:553–563
Gebetsberger J, Wyss L, Mleczko AM, Reuther J, Polacek N (2017) A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress. RNA Biol 14(10):1364–1373
Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F et al (2015) Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351:391–396
Gagnon KT, Corey DR (2012) Argonaute and the nuclear RNAs: new pathways for RNA-mediated control of gene expression. Nucleic Acid Ther 22(1):3–16
Zhang X, He X, Liu C, Liu J, Hu Q, Pan T, Duan X, Liu B, Zhang Y, Chen J, Ma X, Zhang X, Luo H, Zhang H (2016) IL-4 inhibits the biogenesis of an epigenetically suppressive PIWI-interacting RNA to upregulate CD1a molecules on monocytes/dendritic cells. J Immunol 196:1591–1603
Keam SP, Hutvagner G (2015) tRNA-derived fragments (tRFs): emerging new roles for an ancient RNA in the regulation of gene expression. Life (Basel) 5(4):1638–1651
Thompson DM, Lu C, Green PJ, Parker R (2008) tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14(10):2095–2103
Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF, Anderson P (2010) Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem 285(14):10959–10968
Saikia M, Jobava R, Parisien M, Putnam A, Krokowski D, Gao XH, Guan BJ, Yuan Y, Jankowsky E, Feng Z, Hu GF, Pusztai-Carey M, Gorla M, Sepuri NBV, Pan T, Hatzoglou M (2014) Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol Cell Biol 34:2450–2463
Keam SP, Sobala A, Ten Have S, Hutvagner G (2017) tRNA-derived RNA fragments associate with human multisynthetase complex (MSC) and modulate ribosomal protein translation. J Proteome Res 16(2):413–420
Huang B, Yang H, Cheng X, Wang D, Fu S, Shen W, Zhang Q, Zhang L, Xue Z, Li Y, da Y, Yang Q, Li Z, Liu L, Qiao L, Kong Y, Yao Z, Zhao P, Li M, Zhang R (2017) tRF/miR-1280 suppresses stem cell-like cells and metastasis in colorectal cancer. Cancer Res 77(12):3194–3206
Martens-Uzunova ES, Olvedy M, Jenster G (2013) Beyond microRNA - novel RNAs derived from small non-coding RNA and their implication in cancer. Cancer Lett 340(2):201–211
Anderson P, Ivanov P (2014) tRNA fragments in human health and disease.[J]. FEBS Let 588(23):4297–4304
Selitsky SR, Baran-Gale J, Honda M, Yamane D, Masaki T, Fannin EE, Guerra B, Shirasaki T, Shimakami T, Kaneko S, Lanford RE, Lemon SM, Sethupathy P (2015) Small tRNA-derived RNAs are increased and more abundant than microRNAs in chronic hepatitis B and C. Sci Rep 5(1):7675
Wang Q, Lee I, Ren J, Ajay SS, Lee YS, Bao X (2013) Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol Ther 21:368–379
Deng J, Ptashkin RN, Chen Y, Cheng Z, Liu G, Phan T, Deng X, Zhou J, Lee I, Lee YS, Bao X (2015) Respiratory syncytial virus utilizes a tRNA fragment to suppress antiviral responses through a novel targeting mechanism. Mol Ther 23:1622–1629
Mishima E, Inoue C, Saigusa D, Inoue R, Ito K, Suzuki Y, Jinno D, Tsukui Y, Akamatsu Y, Araki M, Araki K, Shimizu R, Shinke H, Suzuki T, Takeuchi Y, Shima H, Akiyama Y, Toyohara T, Suzuki C, Saiki Y, Tominaga T, Miyagi S, Kawagisihi N, Soga T, Ohkubo T, Yamamura K, Imai Y, Masuda S, Sabbisetti V, Ichimura T, Mount DB, Bonventre JV, Ito S, Tomioka Y, Itoh K, Abe T (2014) Conformational change in transfer RNA is an early indicator of acute cellular damage. J Am Soc Nephrol 25(10):2316–2326
Short AK, Yeshurun S, Powell R, Perreau VM, Fox A, Kim JH, Pang TY, Hannan AJ (2017) Exercise alters mouse sperm small noncoding RNAs and induces a transgenerational modification of male offspring conditioned fear and anxiety. Transl Psychiatry 7(5):e1114
Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng GH, Peng H, Zhang X, Zhang Y, Qian J, Duan E, Zhai Q, Zhou Q (2016) Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351(6271):397–400
Greenway MJ, Andersen PM, Russ C, Ennis S, Cashman S, Donaghy C, Patterson V, Swingler R, Kieran D, Prehn J, Morrison KE, Green A, Acharya KR, Brown RH, Hardiman O (2006) ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nat Genet 38:411–413
Van Es MA, Schelhaas HJ, van Vught PW, Ticozzi N, Andersen PM, Groen EJ, Schulte C, Blauw HM, Koppers M, Diekstra FP et al (2013) Angiogenin variants in Parkinson disease and amyotrophic lateral sclerosis. Ann Neurol 70:964–973
Abbasi-Moheb L, Mertel S, Gonsior M, Nouri-Vahid L, Kahrizi K, Cirak S, Wieczorek D, Motazacker MM, Esmaeeli-Nieh S, Cremer K, Weißmann R, Tzschach A, Garshasbi M, Abedini SS, Najmabadi H, Ropers HH, Sigrist SJ, Kuss AW (2012) Mutations in NSUN2 cause autosomal-recessive intellectual disability. Am J Hum Genet 90:847–855
Khan MA, Rafiq MA, Noor A, Hussain S, Flores JV, Rupp V, Vincent AK, Malli R, Ali G, Khan FS, Ishak GE, Doherty D, Weksberg R, Ayub M, Windpassinger C, Ibrahim S, Frye M, Ansar M, Vincent JB (2012) Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am J Hum Genet 90:856–863
Martinez FJ, Lee JH, Lee JE, Blanco S, Nickerson E, Gabriel S, Frye M, Al-Gazali L, Gleeson JG (2012) Whole exome sequencing identifies a splicing mutation in NSUN2 as a cause of a Dubowitz-like syndrome. J Med Genet 49:380–385
Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M et al (2014) Aberrant methylation of tRNAs links cellular stress to neurodevelopmental disorders. EMBO J 17:2020–2039
Uchiumi T, Fotovati A, Sasaguri T, Shibahara K, Shimada T, Fukuda T, Nakamura T, Izumi H, Tsuzuki T, Kuwano M, Kohno K (2006) YB-1 is important for an early stage embryonic development - neural tube formation and cell proliferation. J Biol Chem 281:40440–40449
Okamura K,Lai E C (2008) Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol 9(9):673–678
Cassandrini D, Biancheri R, Tessa A, Di Rocco M, Di Capua M, Bruno C, Denora PS, Sartori S, Rossi A, Nozza P et al (2010) Pontocerebellar hypoplasia clinical, pathologic, and genetic studies. Neurology 75(16):1459–1464
Wu D, Yu W, Kishikawa H, Folkerth RD, Iafrate AJ, Shen Y, Xin W, Sims K, Hu GF (2007) Angiogenin loss-of-function mutations in amyotrophic lateral sclerosis. Ann Neurol 62:609–617
Schopman NC, Heynen S, Haasnoot J, Berkhout B (2010) A miRNA-tRNA mix-up: tRNA origin of proposed miRNA. RNA Biol 7:573–576
Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F (2010) RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev 24:1590–1595
Schorn AJ, Gutbrod MJ, LeBlanc C, Martienssen R (2017) LTR-retrotransposon control by tRNA-derived small RNAs. Cell 170(1):61–71.e11
Vojtech L, Woo S, Hughes S, Levy C, Ballweber L, Sauteraud RP, Strobl J, Westerberg K, Gottardo R, Tewari M, Hladik F (2014) Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res 42:7290–7304
Kim HK, Fuchs G, Wang S, Wei W, Zhang Y, Park H, Roy-Chaudhuri B, Li P, Xu J, Chu K, Zhang F, Chua MS, So S, Zhang QC, Sarnow P, Kay MA (2017) A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature 552(7683):57–62
Guzzi N, Cieśla M, Ngoc PCT, Lang S, Arora S, Dimitriou M, Pimková K, Sommarin MNE, Munita R, Lubas M, Lim Y, Okuyama K, Soneji S, Karlsson G, Hansson J, Jönsson G, Lund AH, Sigvardsson M, Hellström-Lindberg E, Hsieh AC, Bellodi C (2018) Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell 173(5):1204–1216.e26
Lyons SM, Gudanis D, Coyne SM, Gdaniec Z, Ivanov P (2017) Identification of functional tetramolecular RNA G-quadruplexes derived from transfer RNAs. Nat Commun 8(1):1127
Funding
This study was supported by grants from the Applied Research Project on Nonprofit Technology of Zhejiang Province (no. 2016C33177), the Scientific Innovation Team Project of Ningbo (no. 2017C110019), the National Natural Science Foundation of China (no. 81772279), and the K.C. Wong Magna Fund in Ningbo University.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Shen, Y., Yu, X., Zhu, L. et al. Transfer RNA-derived fragments and tRNA halves: biogenesis, biological functions and their roles in diseases. J Mol Med 96, 1167–1176 (2018). https://doi.org/10.1007/s00109-018-1693-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-018-1693-y