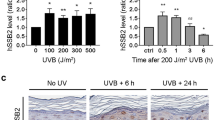
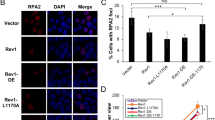
Abstract
In mammalian cells, the bulky DNA adducts caused by ultraviolet radiation are mainly repaired via the nucleotide excision repair (NER) pathway; some defects in this pathway lead to a genetic disorder known as xeroderma pigmentosum (XP). Ribosomal protein S3 (rpS3), a constituent of the 40S ribosomal subunit, is a multi-functional protein with various extra-ribosomal functions, including a role in the cellular stress response and DNA repair-related activities. We report that rpS3 associates with transcription factor IIH (TFIIH) via an interaction with the xeroderma pigmentosum complementation group D (XPD) protein and complements its function in the NER pathway. For optimal repair of UV-induced duplex DNA lesions, the strong helicase activity of the TFIIH complex is required for unwinding damaged DNA around the lesion. Here, we show that XP-D cells overexpressing rpS3 showed markedly increased resistance to UV radiation through XPD and rpS3 interaction. Additionally, the knockdown of rpS3 caused reduced NER efficiency in HeLa cells and the overexpression of rpS3 partially restored helicase activity of the TFIIH complex of XP-D cells in vitro. We also present data suggesting that rpS3 is involved in post-excision processing in NER, assisting TFIIH in expediting the repair process by increasing its turnover rate when DNA is damaged. We propose that rpS3 is an accessory protein of the NER pathway and its recruitment to the repair machinery augments repair efficiency upon UV damage by enhancing XPD helicase function and increasing its turnover rate.







Similar content being viewed by others
Availability of data and material
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- CPD:
-
Cyclobutane pyrimidine dimer
- NER:
-
Nucleotide excision repair
- rpS3:
-
Ribosomal protein S3
- TFIIH:
-
Transcription factor IIH
- XP:
-
Xeroderma pigmentosum
References
Coin F, Oksenych V, Egly J-M (2007) Distinct roles for the XPB/p52 and XPD/p44 Subcomplexes of TFIIH in damaged DNA opening during nucleotide excision repair. Mol Cell 26(2):245–256. https://doi.org/10.1016/j.molcel.2007.03.009
Friedberg ECW, Siede GC, Wood RD, Schultz RA, Ellenberger T (2006) DNA repair and mutagenesis, 2nd edn. ASM press, Washington, USA
Brandsma I, van Gent DC (2012) Pathway choice in DNA double strand break repair: observations of a balancing act. Genome Integr 3(1):9. https://doi.org/10.1186/2041-9414-3-9
Compe E, Egly J-M (2012) TFIIH: when transcription met DNA repair. Nat Rev Mol Cell Biol 13(6):343–354
Egly J-M, Coin F (2011) A history of TFIIH: Two decades of molecular biology on a pivotal transcription/repair factor. DNA Repair 10(7):714–721. https://doi.org/10.1016/j.dnarep.2011.04.021
Kuper J, Kisker C (2012) Damage recognition in nucleotide excision DNA repair. Curr Opin Struct Biol 22(1):88–93. https://doi.org/10.1016/j.sbi.2011.12.002
Drapkin R, Reardon JT, Ansari A, Huang J-C, Zawel L, Ahn K, Sancar A, Reinberg D (1994) Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature 368(6473):769–772. https://doi.org/10.1038/368769a0
Dip R, Camenisch U, Naegeli H (2004) Mechanisms of DNA damage recognition and strand discrimination in human nucleotide excision repair. DNA Repair (Amst) 3(11):1409–1423
Kamileri I, Karakasilioti I, Garinis GA (2012) Nucleotide excision repair: new tricks with old bricks. Trends Genet 28(11):566–573. https://doi.org/10.1016/j.tig.2012.06.004
Zotter A, Luijsterburg MS, Warmerdam DO, Ibrahim S, Nigg A, van Cappellen WA, Hoeijmakers JHJ, van Driel R, Vermeulen W, Houtsmuller AB (2006) Recruitment of the nucleotide excision repair endonuclease XPG to sites of UV-induced DNA damage depends on functional TFIIH. Mol Cell Biol 26(23):8868–8879. https://doi.org/10.1128/mcb.00695-06
Evans E, Moggs JG, Hwang JR, Egly J-M, Wood RD (1997) Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. The EMBO J 16(21):6559–6573. https://doi.org/10.1093/emboj/16.21.6559
Lehmann AR (2003) DNA repair-deficient diseases, xeroderma pigmentosum Cockayne syndrome and trichothiodystrophy. Biochimie 85(11):1101–1111. https://doi.org/10.1016/j.biochi.2003.09.010
Riedl T, Hanaoka F, Egly J-M (2003) The comings and goings of nucleotide excision repair factors on damaged DNA. The EMBO J 22(19):5293–5303. https://doi.org/10.1093/emboj/cdg489
Taylor EM, Broughton BC, Botta E, Stefanini M, Sarasin A, Jaspers NG, Fawcett H, Harcourt SA, Arlett CF, Lehmann AR (1997) Xeroderma pigmentosum and trichothiodystrophy are associated with different mutations in the XPD (ERCC2) repair/transcription gene. Proc Natl Acad Sci USA 94(16):8658–8663
Kim J, Chubatsu LS, Admon A, Stahl J, Fellous R, Linn S (1995) Implication of mammalian ribosomal protein S3 in the processing of DNA damage. J Biol Chem 270(23):13620–13629. https://doi.org/10.1074/jbc.270.23.13620
Kim SH, Lee JY, Kim J (2005) Characterization of a wide range base-damage-endonuclease activity of mammalian rpS3. Biochem Biophys Res Commun 328(4):962–967. https://doi.org/10.1016/j.bbrc.2005.01.045
Jung S-O, Lee JY, Kim J (2001) Yeast ribosomal protein S3 has an endonuclease activity on AP DNA. Mol Cells 12(1):84–90
Yang HW, Jung Y, Kim HD, Kim J (2020) Ribosomal protein S3-derived repair domain peptides regulate UV-induced matrix metalloproteinase-1. Biochem Biophys Res Commun 530(1):149–154. https://doi.org/10.1016/j.bbrc.2020.06.094
Choi SH, Kim SY, An JJ, Lee SH, Kim DW, Ryu HJ, Lee NI, Yeo SI, Jang SH, Won MH, Kang TC, Kwon HJ, Cho SW, Kim J, Lee KS, Park J, Eum WS, Choi SY (2006) Human PEP-1-ribosomal protein S3 protects against UV-induced skin cell death. FEBS Lett 580(30):6755–6762. https://doi.org/10.1016/j.febslet.2006.11.038
Yang HW, Kim HD, Kim J (2019) The DNA repair domain of human rpS3 protects against photoaging by removing cyclobutane pyrimidine dimers. FEBS Lett 593(15):2060–2068. https://doi.org/10.1002/1873-3468.13479
Kim Y, Kim HD (1833) Kim J (2013) Cytoplasmic ribosomal protein S3 (rpS3) plays a pivotal role in mitochondrial DNA damage surveillance. Biochim Biophys Acta 12:2943–2952. https://doi.org/10.1016/j.bbamcr.2013.07.015
Han SH, Chung JH, Kim J, Kim K-S, Han YS (2017) New role of human ribosomal protein S3: Regulation of cell cycle via phosphorylation by cyclin-dependent kinase 2. Oncol Lett 13(5):3681–3687. https://doi.org/10.3892/ol.2017.5906
Jung Y, Kim HD, Yang HW, Kim HJ, Jang C-Y, Kim J (2017) Modulating cellular balance of Rps3 mono-ubiquitination by both Hel2 E3 ligase and Ubp3 deubiquitinase regulates protein quality control. Exp Mol Med 49(11):e390–e390. https://doi.org/10.1038/emm.2017.128
Limoncelli KA, Merrikh CN, Moore MJ (2017) ASC1 and RPS3: new actors in 18S nonfunctional rRNA decay. RNA 23(12):1946–1960. https://doi.org/10.1261/rna.061671.117
Jang CY, Kim HD, Kim J (2012) Ribosomal protein S3 interacts with TRADD to induce apoptosis through caspase dependent JNK activation. Biochem Biophys Res Commun 421(3):474–478. https://doi.org/10.1016/j.bbrc.2012.04.020
Kim SH, Kim J (2006) Reduction of invasion in human fibrosarcoma cells by ribosomal protein S3 in conjunction with Nm23-H1 and ERK. Biochim Biophys Acta 1763(8):823–832. https://doi.org/10.1016/j.bbamcr.2006.03.011
Gao X, Wan F, Mateo K, Callegari E, Wang D, Deng W, Puente J, Li F, Chaussee MS, Finlay BB, Lenardo MJ, Hardwidge PR (2009) Bacterial effector binding to ribosomal protein s3 subverts NF-kappaB function. PLoS Pathog 5(12):e1000708. https://doi.org/10.1371/journal.ppat.1000708
Gao X, Hardwidge PR (2011) Ribosomal protein S3: a multifunctional target of attaching/effacing bacterial pathogens. Frontiers in Microbiology 2:137. https://doi.org/10.3389/fmicb.2011.00137
Kemp MG, Reardon JT, Lindsey-Boltz LA, Sancar A (2012) Mechanism of release and fate of excised oligonucleotides during nucleotide excision repair. J Biol Chem 287(27):22889–22899. https://doi.org/10.1074/jbc.M112.374447
Azqueta A, Langie SAS, Slyskova J, Collins AR (2013) Measurement of DNA base and nucleotide excision repair activities in mammalian cells and tissues using the comet assay—a methodological overview. DNA Repair 12(11):1007–1010. https://doi.org/10.1016/j.dnarep.2013.07.011
Dubaele S, De Santis LP, Bienstock RJ, Keriel A, Stefanini M, Van Houten B, Egly J-M (2003) Basal transcription defect discriminates between xeroderma pigmentosum and trichothiodystrophy in XPD patients. Mol Cell 11(6):1635–1646. https://doi.org/10.1016/S1097-2765(03)00182-5
Katsumi S, Kobayashi N, Imoto K, Nakagawa A, Yamashina Y, Muramatsu T, Shirai T, Miyagawa S, Sugiura S, Hanaoka F, Matsunaga T, Nikaido O, Mori T (2001) In situ visualization of ultraviolet-light-induced dna damage repair in locally irradiated human fibroblasts. J Investig Dermatol 117(5):1156–1161. https://doi.org/10.1046/j.0022-202x.2001.01540.x
Arab HH, Wani G, Ray A, Shah ZI, Zhu Q, Wani AA (2010) Dissociation of CAK from core TFIIH reveals a functional link between XP-G/CS and the TFIIH disassembly state. PLoS ONE 5(6):e11007. https://doi.org/10.1371/journal.pone.0011007
Seroz T, Perez C, Bergmann E, Bradsher J, Egly J-M (2000) p44/SSL1, the regulatory subunit of the XPD/RAD3 helicase, plays a crucial role in the transcriptional activity of TFIIH. J Biol Chem 275(43):33260–33266. https://doi.org/10.1074/jbc.M004764200
Coin F, Bergmann E, Tremeau-Bravard A, Egly J-M (1999) Mutations in XPB and XPD helicases found in xeroderma pigmentosum patients impair the transcription function of TFIIH. EMBO J 18(5):1357–1366. https://doi.org/10.1093/emboj/18.5.1357
Hu J, Choi J-H, Gaddameedhi S, Kemp MG, Reardon JT, Sancar A (2013) Nucleotide excision repair in human cells: fate of the excised oligonucleotide carrying dna damage in vivo. J Biol Chem 288(29):20918–20926. https://doi.org/10.1074/jbc.M113.482257
Sung P, Bailly V, Weber C, Thompson LH, Prakash L, Prakash S (1993) Human xeroderma pigmentosum group D gene encodes a DNA helicase. Nature 365(6449):852–855
Lingelbach K, Dobberstein B (1988) An extended RNA/RNA duplex structure within the coding region of mRNA does not block translational elongation. Nucleic Acids Res 16(8):3405–3414
Takyar S, Hickerson RP, Noller HF (2005) mRNA helicase activity of the ribosome. Cell 120(1):49–58
Oksenych V, de Jesus BB, Zhovmer A, Egly JM, Coin F (2009) Molecular insights into the recruitment of TFIIH to sites of DNA damage. EMBO J 28(19):2971–2980. https://doi.org/10.1038/emboj.2009.230
Flores O, Lu H, Reinberg D (1992) Factors involved in specific transcription by mammalian RNA polymerase II. Identification and characterization of factor IIH. J Biol Chem 267(4):2786–2793
Ahn EH, Kim DW, Kang HW, Shin MJ, Won MH, Kim J, Kim DJ, Kwon O-S, Kang T-C, Han KH, Park J, Eum WS, Choi SY (2010) Transduced PEP-1-ribosomal protein S3 (rpS3) ameliorates 12-O-tetradecanoylphorbol-13-acetate-induced inflammation in mice. Toxicology 276(3):192–197. https://doi.org/10.1016/j.tox.2010.08.004
Singh NP, McCoy MT, Tice RR, Schneider EL (1988) A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res 175(1):184–191. https://doi.org/10.1016/0014-4827(88)90265-0
Funding
This paper was supported by National Research Foundation of Korea Grant NRF-2020R1A2C2100803, NRF-2019S1A5A2A03050121 and Korea University Grant.
Author information
Authors and Affiliations
Contributions
YJP, SHK, and JK designed research; YJP, SHK, SML, BSC, and CIS performed research; YJP, SHK, H-DK, and T-SK contributed to data analysis and research design; H-DK and JK supervised the research; and YJP and JK wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Park, Y.J., Kim, S.H., Kim, T.S. et al. Ribosomal protein S3 associates with the TFIIH complex and positively regulates nucleotide excision repair. Cell. Mol. Life Sci. 78, 3591–3606 (2021). https://doi.org/10.1007/s00018-020-03754-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-020-03754-x