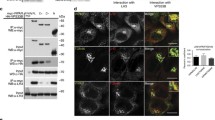
Abstract
The P3H1/CRTAP/PPIB complex is essential for prolyl 3-hydroxylation and folding of procollagens in the endoplasmic reticulum (ER). Deficiency in any component of this ternary complex is associated with the misfolding of collagen and the onset of osteogenesis imperfecta. However, little structure information is available about how this ternary complex is assembled and retained in the ER. Here, we assessed the role of the KDEL sequence of P3H1 and probed the spatial interactions of PPIB in the complex. We show that the KDEL sequence is essential for retaining the P3H1 complex in the ER. Its removal resulted in co-secretion of P3H1 and CRTAP out of the cell, which was mediated by the binding of P3H1 N-terminal domain with CRTAP. The secreted P3H1/CRTAP can readily bind PPIB with their C-termini close to PPIB in the ternary complex. Cysteine modification, crosslinking, and mass spectrometry experiments identified PPIB surface residues involved in the complex formation, and showed that the surface of PPIB is extensively covered by the binding of P3H1 and CRTAP. Most importantly, we demonstrated that one disease-associated pathological PPIB mutation on the binding interface did not affect the PPIB prolyl-isomerase activity, but disrupted the formation of P3H1/CRTAP/PPIB ternary complex. This suggests that defects in the integrity of the P3H1 ternary complex are associated with pathological collagen misfolding. Taken together, these results provide novel structural information on how PPIB interacts with other components of the P3H1 complex and indicate that the integrity of P3H1 complex is required for proper collagen formation.







Similar content being viewed by others
Abbreviations
- ER:
-
Endoplasmic reticulum
- P3H1:
-
Prolyl 3-hydroxylase 1
- CRTAP:
-
Cartilage-associated protein
- PPIB:
-
Peptidyl–prolyl cis–trans isomerase B
- OI:
-
Osteogenesis imperfecta
- mPEG:
-
NEM-PEG2000
- MW:
-
Molecular weight
- HC:
-
Hyperelastosis cutis
- WT:
-
Wild type
- IPTG:
-
Isopropyl-β-d-thiogalactopyranoside
- DTT:
-
Dithiothreitol
References
Bächinger HP, Mizuno K, Vranka JA, Boudko SP (2010) Collagen formation and structure. In: Mander L, Liu HW (eds) Comprehensive natural products II: chemistry and biology. Elsevier, Oxford, pp 469–530
Hamaia S, Farndale RW (2014) Integrin recognition motifs in the human collagens. Adv Exp Med Biol 819:127–142
Myllyharju J (2005) Intracellular post-translational modifications of collagens. Top Curr Chem 247:115–147
Myllyharju J, Kivirikko KI (2004) Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet 20(1):33–43. https://doi.org/10.1016/j.tig.2003.11.004
Lamande SR, Bateman JF (1999) Procollagen folding and assembly: the role of endoplasmic reticulum enzymes and molecular chaperones. Semin Cell Dev Biol 10(5):455–464
Jimenez S, Harsch M, Rosenbloom J (1973) Hydroxyproline stabilizes the triple helix of chick tendon collagen. Biochem Biophys Res Commun 52(1):106
Berg RA, Prockop DJ (1973) The thermal transition of a non-hydroxylated form of collagen. Evidence for a role for hydroxyproline in stabilizing the triple-helix of collagen. Biochem Biophys Res Commun 52(1):115
Myllyharju J (2003) Prolyl 4-hydroxylases, the key enzymes of collagen biosynthesis. Matrix Biol 22(1):15–24
Lees JF, Tasab M, Bulleid NJ (1997) Identification of the molecular recognition sequence which determines the type-specific assembly of procollagen. EMBO J 16(5):908
Doege KJ, Fessler JH (1986) Folding of carboxyl domain and assembly of procollagen I. J Biol Chem 261(19):8924–8935
Hammond C, Helenius A (1995) Quality control in the secretory pathway. Curr Opin Cell Biol 7(4):523
Vranka JA, Sakai LY, Bachinger HP (2004) Prolyl 3-hydroxylase 1, enzyme characterization and identification of a novel family of enzymes. J Biol Chem 279(22):23615–23621. https://doi.org/10.1074/jbc.M312807200
Kellokumpu S, Sormunen R, Heikkinen J, Myllylã R (1994) Lysyl hydroxylase, a collagen processing enzyme, exemplifies a novel class of luminally-oriented peripheral membrane proteins in the endoplasmic reticulum. J Biol Chem 269(48):30524–30529
Steinmann B, Bruckner P, Superti-Furga A (1991) Cyclosporin A slows collagen triple-helix formation in vivo: indirect evidence for a physiologic role of peptidyl-prolyl cis–trans-isomerase. J Biol Chem 266(2):1299–1303
Forlino A, Cabral WA, Barnes AM, Marini JC (2011) New perspectives on osteogenesis imperfecta. Nat Rev Endocrinol 7(9):540–557. https://doi.org/10.1038/nrendo.2011.81
Marini JC, Forlino A, Bächinger HP, Bishop NJ, Byers PH, Paepe AD, Fassier F, Fratzl-Zelman N, Kozloff KM, Krakow D, Montpetit K, Semler O (2017) Osteogenesis imperfecta. Nat Rev Dis Prim 3:17052. https://doi.org/10.1038/nrdp.2017.52
Forlino A, Marini JC (2016) Osteogenesis imperfecta. Lancet 387(10028):1657–1671
Alanay Y, Avaygan H, Camacho N, Utine GE, Boduroglu K, Aktas D, Alikasifoglu M, Tuncbilek E, Orhan D, Bakar FT (2010) Mutations in the gene encoding the RER protein FKBP65 cause autosomal-recessive osteogenesis imperfecta. Am J Hum Genet 86(4):551–559
Christiansen HE, Schwarze U, Pyott SM, Alswaid A, Balwi MA, Alrasheed S, Pepin MG, Weis MA, Eyre DR, Byers PH (2010) Homozygosity for a missense mutation in SERPINH1, which encodes the collagen chaperone protein HSP47, results in severe recessive osteogenesis imperfecta. Am J Hum Genet 86(3):389–398
Ito S, Nagata K (2017) Biology of Hsp47 (Serpin H1), a collagen-specific molecular chaperone. Semin Cell Dev Biol 62:142–151
Marini JC, Reich A, Smith SM (2014) Osteogenesis imperfecta due to mutations in non-collagenous genes: lessons in the biology of bone formation. Curr Opin Pediatr 26(4):500–507. https://doi.org/10.1097/MOP.0000000000000117
Morello R, Bertin TK, Chen Y, Hicks J, Tonachini L, Monticone M, Castagnola P, Rauch F, Glorieux FH, Vranka J, Bachinger HP, Pace JM, Schwarze U, Byers PH, Weis M, Fernandes RJ, Eyre DR, Yao Z, Boyce BF, Lee B (2006) CRTAP is required for prolyl 3- hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell 127(2):291–304. https://doi.org/10.1016/j.cell.2006.08.039
Vranka JA, Pokidysheva E, Hayashi L, Zientek K, Mizuno K, Ishikawa Y, Maddox K, Tufa S, Keene DR, Klein R (2010) Prolyl 3-hydroxylase 1 null mice display abnormalities in fibrillar collagen-rich tissues such as tendons, skin and bones. J Biol Chem JBC M110:102228
Choi JW, Sutor SL, Lindquist L, Evans GL, Madden BJ, Bergen HR 3rd, Hefferan TE, Yaszemski MJ, Bram RJ (2009) Severe osteogenesis imperfecta in cyclophilin B-deficient mice. PLoS Genet 5(12):e1000750. https://doi.org/10.1371/journal.pgen.1000750
Marini JC, Cabral WA, Barnes AM, Chang W (2007) Components of the collagen prolyl 3-hydroxylation complex are crucial for normal bone development. Cell Cycle 6(14):1675–1681. https://doi.org/10.4161/cc.6.14.4474
Tonachini L, Morello R, Monticone M, Skaug J, Scherer SW, Cancedda R, Castagnola P (1999) cDNA cloning, characterization and chromosome mapping of the gene encoding human cartilage associated protein (CRTAP). Cytogenet Genome Res 87(3–4):191
Ishikawa Y, Bachinger HP (2013) An additional function of the rough endoplasmic reticulum protein complex prolyl 3-hydroxylase 1. Cartilage-associated protein–cyclophilin B: the CXXXC motif reveals disulfide isomerase activity in vitro. J Biol Chem 288(44):31437–31446. https://doi.org/10.1074/jbc.m113.498063
Chang W, Barnes AM, Cabral WA, Bodurtha JN, Marini JC (2010) Prolyl 3-hydroxylase 1 and CRTAP are mutually stabilizing in the endoplasmic reticulum collagen prolyl 3-hydroxylation complex. Hum Mol Genet 19(2):223–234. https://doi.org/10.1093/hmg/ddp481
Takagi M, Ishii T, Barnes AM, Weis M, Amano N, Tanaka M, Fukuzawa R, Nishimura G, Eyre DR, Marini JC, Hasegawa T (2012) A novel mutation in LEPRE1 that eliminates only the KDEL ER- retrieval sequence causes non-lethal osteogenesis imperfecta. PLoS One 7(5):e36809. https://doi.org/10.1371/journal.pone.0036809
Barbirato C, Trancozo M, Almeida MG, Almeida LS, Santos TO, Duarte JC, Reboucas MR, Sipolatti V, Nunes VR, Paula F (2015) Mutational characterization of the P3H1/CRTAP/CypB complex in recessive osteogenesis imperfecta. Genet Mol Res 14(4):15848–15858. https://doi.org/10.4238/2015.december.1.36
Wisniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6(5):359–362. https://doi.org/10.1038/nmeth.1322
Yang B, Wu YJ, Zhu M, Fan SB, Lin J, Zhang K, Li S, Chi H, Li YX, Chen HF, Luo SK, Ding YH, Wang LH, Hao Z, Xiu LY, Chen S, Ye K, He SM, Dong MQ (2012) Identification of cross-linked peptides from complex samples. Nat Methods 9(9):904–906. https://doi.org/10.1038/nmeth.2099
Kofron JL, Kuzmic P, Kishore V, Colonbonilla E, Rich DH (1991) Determination of kinetic constants for peptidyl prolyl cis–trans isomerases by an improved spectrophotometric assay. Biochemistry 30(25):6127–6134
Dimou M, Venieraki A, Liakopoulos G, Kouri ED, Tampakaki A, Katinakis P (2011) Gene expression and biochemical characterization of Azotobacter vinelandii cyclophilins and protein interaction studies of the cytoplasmic isoform with dnaK and lpxH. J Mol Microbiol Biotechnol 20(3):176–190
Galat A, Bouet F (1994) Cyclophilin-B is an abundant protein whose conformation is similar to cyclophilin-A. FEBS Lett 347(1):31
Mikol V, Kallen J, Walkinshaw MD (1994) X-Ray structure of a cyclophilin B/cyclosporin complex: comparison with cyclophilin A and delineation of its calcineurin-binding domain. Proc Natl Acad Sci USA 91(11):5183–5186
Ishikawa Y, Vranka JA, Boudko SP, Pokidysheva E, Mizuno K, Zientek K, Keene DR, Rashmirraven AM, Nagata K, Winand NJ (2012) Mutation in cyclophilin B that causes hyperelastosis cutis in american quarter horse does not affect peptidylprolyl cis–trans isomerase activity but shows altered cyclophilin B–protein interactions and affects collagen folding. J Biol Chem 287(26):22253–22265
Jiang Y, Pan J, Guo D, Zhang W, Xie J, Fang Z, Guo C, Fang Q, Jiang W, Guo Y (2017) Two novel mutations in the PPIB gene cause a rare pedigree of osteogenesis imperfecta type IX. Clinica Chimica Acta 469:111–118
Price ER, Zydowsky LD, Jin MJ, Baker CH, Mckeon FD, Walsh CT (1991) Human cyclophilin B: a second cyclophilin gene encodes a peptidyl-prolyl isomerase with a signal sequence. Proc Natl Acad Sci USA 88(5):1903–1907
Barnes AM, Chang W, Morello R, Cabral WA, Weis M, Eyre DR, Leikin S, Makareeva E, Kuznetsova N, Uveges TE (2006) Deficiency of cartilage-associated protein in recessive lethal osteogenesis imperfecta. N Engl J Med 355(26):2757–2764
Cabral WA, Chang W, Barnes AM, Weis M, Scott MA, Leikin S, Makareeva E, Kuznetsova NV, Rosenbaum KN, Tifft CJ, Bulas DI, Kozma C, Smith PA, Eyre DR, Marini JC (2007) Prolyl 3-hydroxylase 1 deficiency causes a recessive metabolic bone disorder resembling lethal/severe osteogenesis imperfecta. Nat Genet 39(3):359–365. https://doi.org/10.1038/ng1968
Dijk FSV, Nesbitt IM, Zwikstra EH, Nikkels PGJ, Piersma SR, Fratantoni SA, Jimenez CR, Huizer M, Morsman AC, Cobben JM (2009) PPIB mutations cause severe osteogenesis imperfecta. Am J Hum Genet 85(4):521–527
Baldridge D, Schwarze U, Morello R, Lennington J, Bertin TK, Pace JM, Pepin MG, Weis M, Eyre DR, Walsh J, Lambert D, Green A, Robinson H, Michelson M, Houge G, Lindman C, Martin J, Ward J, Lemyre E, Mitchell JJ, Krakow D, Rimoin DL, Cohn DH, Byers PH, Lee B (2008) CRTAP and LEPRE1 mutations in recessive osteogenesis imperfecta. Hum Mutat 29(12):1435–1442. https://doi.org/10.1002/humu.20799
Ishikawa Y, Wirz J, Vranka JA, Nagata K, Bachinger HP (2009) Biochemical characterization of the prolyl 3-hydroxylase 1. Cartilage-associated protein–cyclophilin B complex. J Biol Chem 284(26):17641–17647. https://doi.org/10.1074/jbc.m109.007070
Homan EP, Lietman C, Grafe I, Lennington J, Morello R, Napierala D, Jiang MM, Munivez EM, Dawson B, Bertin TK, Chen Y, Lua R, Lichtarge O, Hicks J, Weis MA, Eyre D, Lee BH (2014) Differential effects of collagen prolyl 3-hydroxylation on skeletal tissues. PLoS Genet 10(1):e1004121. https://doi.org/10.1371/journal.pgen.1004121
Barnes AM, Carter EM, Cabral WA, Weis M, Chang W, Makareeva E, Leikin S, Rotimi CN, Eyre DR, Raggio CL, Marini JC (2010) Lack of cyclophilin B in osteogenesis imperfecta with normal collagen folding. N Engl J Med 362(6):521–528. https://doi.org/10.1056/NEJMoa0907705
Pyott SM, Schwarze U, Christiansen HE, Pepin MG, Leistritz DF, Dineen R, Harris C, Burton BK, Angle B, Kim K, Sussman MD, Weis M, Eyre DR, Russell DW, McCarthy KJ, Steiner RD, Byers PH (2011) Mutations in PPIB (cyclophilin B) delay type I procollagen chain association and result in perinatal lethal to moderate osteogenesis imperfecta phenotypes. Hum Mol Genet 20(8):1595–1609. https://doi.org/10.1093/hmg/ddr037
Kozlov G, Bastos-Aristizabal S, Maattanen P, Rosenauer A, Zheng F, Killikelly A, Trempe JF, Thomas DY, Gehring K (2010) Structural basis of cyclophilin B binding by the calnexin/calreticulin P-domain. J Biol Chem 285(46):35551–35557. https://doi.org/10.1074/jbc.M110.160101
Acknowledgements
This work was supported in part by Grants from the National Natural Science Foundation of China (81870309, 31570824, and 81572090), the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning, Shanghai PuJiang Program, and Innovation Program of Shanghai Municipal Education Commission (no. 12ZZ113).
Funding
This work was supported in part by Grants from the National Natural Science Foundation of China (81870309, 31570824, and 81572090), the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning, Shanghai PuJiang Program, and Innovation Program of Shanghai Municipal Education Commission (no. 12ZZ113).
Author information
Authors and Affiliations
Contributions
JW and WZ prepared all the P3H1/CRTAP/PPIB mutants. WZ, ZS, JZ, and LF performed the in vitro experiment about the interaction of P3H1 and CRTAP. J.W carried out the PPIB cysteine modification and crosslinking experiment. XL helped to establish the mass spectrometry method. JW and AZ wrote the paper. WY, NZ, and AZ conceived and designed all the experiments.
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wu, J., Zhang, W., Xia, L. et al. Characterization of PPIB interaction in the P3H1 ternary complex and implications for its pathological mutations. Cell. Mol. Life Sci. 76, 3899–3914 (2019). https://doi.org/10.1007/s00018-019-03102-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-019-03102-8