Abstract
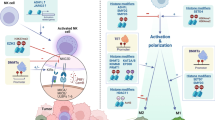
The recent impact of cancer immunotherapies has firmly established the ability and importance of the immune system to fight malignancies. However, the intimate interaction between the highly dynamic tumor and immune cells leads to a selection process driven by genetic and epigenetic processes. As the molecular pathways of cancer resistance mechanisms to immunotherapy become increasingly known, novel therapeutic targets are being tested in combination with immune-stimulating approaches. We here review recent insights into the molecular mechanisms of tumor resistance with particular emphasis on epigenetic processes and place these in the context of previous models.



Similar content being viewed by others
References
Coley WB (1893) The treatment of malignant tumors by repeated inoculations of erysipelas: with a report of ten original cases. Am J Med Sci 105:487–511
McCarthy EF (2006) The toxins of William B. Coley and the treatment of bone and soft-tissue sarcomas. Iowa Orthop J 26:154–158
Hodi FS et al (2010) Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 363(8):711–723
Garon EB et al (2015) Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 372(21):2018–2028
Larkin J, Hodi FS, Wolchok JD (2015) Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med 373(13):1270–1271
Ribas A et al (2016) Association of pembrolizumab with tumor response and survival among patients with advanced melanoma. JAMA 315(15):1600–1609
Robert C et al (2015) Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med 372(26):2521–2532
Sharma P et al (2017) Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168(4):707–723
Anagnostou V et al (2017) Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov 7(3):264–276
Matsushita H et al (2012) Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature 482(7385):400–404
Schreiber RD, Old LJ, Smyth MJ (2011) Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science 331(6024):1565–1570
Verdegaal EM et al (2016) Neoantigen landscape dynamics during human melanoma–T cell interactions. Nature 536(7614):91–95
Spranger S, Bao R, Gajewski TF (2015) Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature 523(7559):231–235
Peng W et al (2016) Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov 6(2):202–216
Shin DS et al (2017) Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov 7(2):188–201
Zaretsky JM et al (2016) Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med 375(9):819–829
Gao J et al (2016) Loss of IFN-gamma pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell 167(2):397 e9–404 e9
Patel SJ et al (2017) Identification of essential genes for cancer immunotherapy. Nature 548(7669):537–542
Spranger S et al (2017) Tumor-residing Batf3 dendritic cells are required for effector T cell trafficking and adoptive T cell therapy. Cancer Cell 31(5):711.e4–723.e4
Casey SC et al (2016) MYC regulates the antitumor immune response through CD47 and PD-L1. Science 352(6282):227–231
Lastwika KJ et al (2016) Control of PD-L1 expression by oncogenic activation of the AKT–mTOR pathway in non-small cell lung cancer. Cancer Res 76(2):227–238
Topalian SL, Drake CG, Pardoll DM (2015) Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 27(4):450–461
Spranger S et al (2013) Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med 5(200):200ra116
Benci JL et al (2016) Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell 167(6):1540.e12–1554.e12
Shayan G et al (2017) Adaptive resistance to anti-PD1 therapy by Tim-3 upregulation is mediated by the PI3K–Akt pathway in head and neck cancer. Oncoimmunology 6(1):e1261779
Gao J et al (2017) VISTA is an inhibitory immune checkpoint that is increased after ipilimumab therapy in patients with prostate cancer. Nat Med 23(5):551–555
Brand A et al (2016) LDHA-associated lactic acid production blunts tumor immunosurveillance by T and NK cells. Cell Metab 24(5):657–671
Chaudhary B, Elkord E (2016) Regulatory T cells in the tumor microenvironment and cancer progression: role and therapeutic targeting.Vaccines (Basel). 4(3). pii: E28. https://doi.org/10.1016/j.cellimm.2018.02.008
Lin YC et al (2013) Activated but not resting regulatory T cells accumulated in tumor microenvironment and correlated with tumor progression in patients with colorectal cancer. Int J Cancer 132(6):1341–1350
Yang L et al (2008) Abrogation of TGF beta signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell 13(1):23–35
Solito S et al (2011) A human promyelocytic-like population is responsible for the immune suppression mediated by myeloid-derived suppressor cells. Blood 118(8):2254–2265
Yang L et al (2004) Expansion of myeloid immune suppressor Gr+CD11b+ cells in tumor-bearing host directly promotes tumor angiogenesis. Cancer Cell 6(4):409–421
Kiss M et al (2018) Myeloid cell heterogeneity in cancer: not a single cell alike. Cell Immunol. https://doi.org/10.1016/j.cellimm.2018.02.008
Talmadge JE, Gabrilovich DI (2013) History of myeloid-derived suppressor cells. Nat Rev Cancer 13(10):739–752
Lewis CE, Harney AS, Pollard JW (2016) The multifaceted role of perivascular macrophages in tumors. Cancer Cell 30(2):365
Mantovani A et al (2017) Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol 14(7):399–416
Motz GT et al (2014) Tumor endothelium FasL establishes a selective immune barrier promoting tolerance in tumors. Nat Med 20(6):607–615
Caruana I et al (2015) Heparanase promotes tumor infiltration and antitumor activity of CAR-redirected T lymphocytes. Nat Med 21(5):524–529
Vetizou M et al (2015) Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350(6264):1079–1084
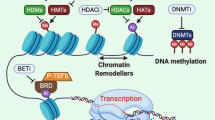
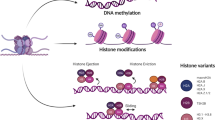
Maio M et al (2015) Molecular pathways: at the crossroads of cancer epigenetics and immunotherapy. Clin Cancer Res 21(18):4040–4047
Kanwal R, Gupta K, Gupta S (2015) Cancer epigenetics: an introduction. Methods Mol Biol 1238:3–25
Nebbioso A et al (2012) Trials with ‘epigenetic’ drugs: an update. Mol Oncol 6(6):657–682
Coulie PG et al (2014) Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer 14(2):135–146
Heninger E, Krueger TE, Lang JM (2015) Augmenting antitumor immune responses with epigenetic modifying agents. Front Immunol 6:29
Huijbers IJ et al (2012) Minimal tolerance to a tumor antigen encoded by a cancer-germline gene. J Immunol 188(1):111–121
Scanlan MJ et al (2002) Cancer/testis antigens: an expanding family of targets for cancer immunotherapy. Immunol Rev 188:22–32
James SR, Link PA, Karpf AR (2006) Epigenetic regulation of X-linked cancer/germline antigen genes by DNMT1 and DNMT3b. Oncogene 25(52):6975–6985
Yu J et al (2004) Methylation profiles of thirty four promoter-CpG islands and concordant methylation behaviours of sixteen genes that may contribute to carcinogenesis of astrocytoma. BMC Cancer 4:65
Yu J et al (2002) Methylation profiling of twenty promoter-CpG islands of genes which may contribute to hepatocellular carcinogenesis. BMC Cancer 2:29
Smith HA et al (2011) Expression and immunotherapeutic targeting of the SSX family of cancer-testis antigens in prostate cancer. Cancer Res 71(21):6785–6795
Weber J et al (1994) Expression of the MAGE-1 tumor antigen is up-regulated by the demethylating agent 5-aza-2′-deoxycytidine. Cancer Res 54(7):1766–1771
Dubovsky JA, McNeel DG (2007) Inducible expression of a prostate cancer-testis antigen, SSX-2, following treatment with a DNA methylation inhibitor. Prostate 67(16):1781–1790
Goodyear O et al (2010) Induction of a CD8+ T-cell response to the MAGE cancer testis antigen by combined treatment with azacitidine and sodium valproate in patients with acute myeloid leukemia and myelodysplasia. Blood 116(11):1908–1918
Rao M et al (2011) Inhibition of histone lysine methylation enhances cancer-testis antigen expression in lung cancer cells: implications for adoptive immunotherapy of cancer. Cancer Res 71(12):4192–4204
Toor AA et al (2012) Epigenetic induction of adaptive immune response in multiple myeloma: sequential azacitidine and lenalidomide generate cancer testis antigen-specific cellular immunity. Br J Haematol 158(6):700–711
Garcia-Lora A et al (2003) MHC class I-deficient metastatic tumor variants immunoselected by T lymphocytes originate from the coordinated downregulation of APM components. Int J Cancer 106(4):521–527
Serrano A et al (2001) Rexpression of HLA class I antigens and restoration of antigen-specific CTL response in melanoma cells following 5-aza-2′-deoxycytidine treatment. Int J Cancer 94(2):243–251
Li H et al (2014) Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget 5(3):587–598
Setiadi AF et al (2007) Epigenetic control of the immune escape mechanisms in malignant carcinomas. Mol Cell Biol 27(22):7886–7894
Setiadi AF et al (2008) Epigenetic enhancement of antigen processing and presentation promotes immune recognition of tumors. Cancer Res 68(23):9601–9607
Ritter C et al (2017) Epigenetic priming restores the HLA class-I antigen processing machinery expression in Merkel cell carcinoma. Sci Rep 7(1):2290
Chou SD et al (2005) Histone acetylation regulates the cell type specific CIITA promoters, MHC class II expression and antigen presentation in tumor cells. Int Immunol 17(11):1483–1494
Khan AN, Magner WJ, Tomasi TB (2004) An epigenetically altered tumor cell vaccine. Cancer Immunol Immunother 53(8):748–754
Magner WJ et al (2000) Activation of MHC class I, II, and CD40 gene expression by histone deacetylase inhibitors. J Immunol 165(12):7017–7024
Maeda T et al (2000) Up-regulation of costimulatory/adhesion molecules by histone deacetylase inhibitors in acute myeloid leukemia cells. Blood 96(12):3847–3856
Nencioni A et al (2007) Histone deacetylase inhibitors affect dendritic cell differentiation and immunogenicity. Clin Cancer Res 13(13):3933–3941
Rosenzweig JM et al (2013) KLF4 modulates expression of IL-6 in dendritic cells via both promoter activation and epigenetic modification. J Biol Chem 288(33):23868–23874
Hervouet E et al (2013) DNA methylation and apoptosis resistance in cancer cells. Cells 2(3):545–573
Hopkins-Donaldson S et al (2003) Silencing of death receptor and caspase-8 expression in small cell lung carcinoma cell lines and tumors by DNA methylation. Cell Death Differ 10(3):356–364
Eramo A et al (2005) Inhibition of DNA methylation sensitizes glioblastoma for tumor necrosis factor-related apoptosis-inducing ligand-mediated destruction. Cancer Res 65(24):11469–11477
Lucas DM et al (2004) The histone deacetylase inhibitor MS-275 induces caspase-dependent apoptosis in B-cell chronic lymphocytic leukemia cells. Leukemia 18(7):1207–1214
Insinga A et al (2005) Inhibitors of histone deacetylases induce tumor-selective apoptosis through activation of the death receptor pathway. Nat Med 11(1):71–76
Natoni F et al (2005) Sodium butyrate sensitises human pancreatic cancer cells to both the intrinsic and the extrinsic apoptotic pathways. Biochim Biophys Acta 1745(3):318–329
Lee PP et al (2001) A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15(5):763–774
de Araújo-Souza PS, Hanschke SCH, Viola JPB (2015) Epigenetic control of interferon-gamma expression in CD8 T cells. J Immunol Res 2015:849573
Harland KL et al (2014) Epigenetic plasticity of Cd8a locus during CD8(+) T-cell development and effector differentiation and reprogramming. Nat Commun 5:3547
Wherry EJ, Kurachi M (2015) Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 15(8):486–499
Utzschneider DT et al (2013) T cells maintain an exhausted phenotype after antigen withdrawal and population reexpansion. Nat Immunol 14(6):603–610
Ahn E et al (2016) Demethylation of the PD-1 promoter is imprinted during the effector phase of CD8 T cell exhaustion. J Virol 90(19):8934–8946
Youngblood B et al (2013) Cutting edge: prolonged exposure to HIV reinforces a poised epigenetic program for PD-1 expression in virus-specific CD8 T cells. J Immunol 191(2):540–544
Youngblood B et al (2011) Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8(+) T cells. Immunity 35(3):400–412
Sen DR et al (2016) The epigenetic landscape of T cell exhaustion. Science 354(6316):1165–1169
Raeber ME et al (2018) The role of cytokines in T-cell memory in health and disease. Immunol Rev 283(1):176–193
Pauken KE et al (2016) Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354(6316):1160–1165
Topper MJ et al (2017) Epigenetic therapy ties MYC depletion to reversing immune evasion and treating lung cancer. Cell 171(6):1284.e21–1300.e21
Ghoneim HE et al (2017) De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell 170(1):142 e19–157 e19
Wrangle J et al (2013) Alterations of immune response of non-small cell lung cancer with azacytidine. Oncotarget 4(11):2067–2079
Yang H et al (2014) Expression of PD-L1, PD-L2, PD-1 and CTLA4 in myelodysplastic syndromes is enhanced by treatment with hypomethylating agents. Leukemia 28(6):1280–1288
Boyman O, Sprent J (2012) The role of interleukin-2 during homeostasis and activation of the immune system. Nat Rev Immunol 12(3):180–190
Wing K, Yamaguchi T, Sakaguchi S (2011) Cell-autonomous and -non-autonomous roles of CTLA-4 in immune regulation. Trends Immunol 32(9):428–433
Letourneau S et al (2009) IL-2- and CD25-dependent immunoregulatory mechanisms in the homeostasis of T-cell subsets. J Allergy Clin Immunol 123(4):758–762
Ohkura N, Kitagawa Y, Sakaguchi S (2013) Development and maintenance of regulatory T cells. Immunity 38(3):414–423
Overacre AE, Vignali DA (2016) T(reg) stability: to be or not to be. Curr Opin Immunol 39:39–43
Ohkura N et al (2012) T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity 37(5):785–799
Lal G et al (2009) Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J Immunol 182(1):259–273
Polansky JK et al (2008) DNA methylation controls Foxp3 gene expression. Eur J Immunol 38(6):1654–1663
Sahakian E et al (2015) Histone deacetylase 11: a novel epigenetic regulator of myeloid derived suppressor cell expansion and function. Mol Immunol 63(2):579–585
Kim K et al (2014) Eradication of metastatic mouse cancers resistant to immune checkpoint blockade by suppression of myeloid-derived cells. Proc Natl Acad Sci USA 111(32):11774–11779
Youn JI et al (2013) Epigenetic silencing of retinoblastoma gene regulates pathologic differentiation of myeloid cells in cancer. Nat Immunol 14(3):211–220
Franklin RA et al (2014) The cellular and molecular origin of tumor-associated macrophages. Science 344(6186):921–925
Pollard JW (2004) Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer 4(1):71–78
Sica A et al (2006) Tumour-associated macrophages are a distinct M2 polarised population promoting tumour progression: potential targets of anti-cancer therapy. Eur J Cancer 42(6):717–727
Ivashkiv LB (2013) Epigenetic regulation of macrophage polarization and function. Trends Immunol 34(5):216–223
Ishii M et al (2009) Epigenetic regulation of the alternatively activated macrophage phenotype. Blood 114(15):3244–3254
Heeb LE et al (2018) Regulation of neutrophils in type 2 immune responses. Curr Opin Immunol 54:115–122
Chang YC et al (2008) Epigenetic control of MHC class II expression in tumor-associated macrophages by decoy receptor 3. Blood 111(10):5054–5063
Margueron R, Reinberg D (2011) The Polycomb complex PRC2 and its mark in life. Nature 469(7330):343–349
Comet I et al (2016) Maintaining cell identity: pRC2-mediated regulation of transcription and cancer. Nat Rev Cancer 16(12):803–810
Kim KH, Roberts CW (2016) Targeting EZH2 in cancer. Nat Med 22(2):128–134
Christofides A et al (2016) Epigenetic regulation of cancer biology and anti-tumor immunity by EZH2. Oncotarget 7(51):85624–85640
Holling TM et al (2007) A role for EZH2 in silencing of IFN-gamma inducible MHC2TA transcription in uveal melanoma. J Immunol 179(8):5317–5325
McCabe MT et al (2012) EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492(7427):108–112
Verma SK et al (2012) Identification of potent, selective, cell-active inhibitors of the histone lysine methyltransferase EZH2. ACS Med Chem Lett 3(12):1091–1096
van Vlerken LE et al (2013) EZH2 is required for breast and pancreatic cancer stem cell maintenance and can be used as a functional cancer stem cell reporter. Stem Cells Transl Med 2(1):43–52
Adhikary G et al (2015) Survival of skin cancer stem cells requires the Ezh2 polycomb group protein. Carcinogenesis 36(7):800–810
Zingg D et al (2015) The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat Commun 6:6051
Cao Q et al (2008) Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene 27(58):7274–7284
Ma DN et al (2016) MicroRNA-26a suppresses epithelial-mesenchymal transition in human hepatocellular carcinoma by repressing enhancer of zeste homolog 2. J Hematol Oncol 9:1
Mikucki ME et al (2015) Non-redundant requirement for CXCR3 signalling during tumoricidal T-cell trafficking across tumour vascular checkpoints. Nat Commun 6:7458
Peng D et al (2015) Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature 527(7577):249–253
Nagarsheth N et al (2016) PRC2 epigenetically silences Th1-type chemokines to suppress effector T-cell trafficking in colon cancer. Cancer Res 76(2):275–282
Zingg D et al (2017) The histone methyltransferase Ezh2 controls mechanisms of adaptive resistance to tumor immunotherapy. Cell Rep 20(4):854–867
Arenas-Ramirez N, Woytschak J, Boyman O (2015) Interleukin-2: biology, design and application. Trends Immunol 36(12):763–777
Arenas-Ramirez N et al (2016) Improved cancer immunotherapy by a CD25-mimobody conferring selectivity to human interleukin-2. Sci Transl Med 8(367):367ra166
Landsberg J et al (2012) Melanomas resist T-cell therapy through inflammation-induced reversible dedifferentiation. Nature 490(7420):412–416
Pan D et al (2018) A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science 359(6377):770–775
Tumes DJ et al (2013) The polycomb protein Ezh2 regulates differentiation and plasticity of CD4(+) T helper type 1 and type 2 cells. Immunity 39(5):819–832
Yang XP et al (2015) EZH2 is crucial for both differentiation of regulatory T cells and T effector cell expansion. Sci Rep 5:10643
Tong Q et al (2014) Ezh2 regulates transcriptional and posttranslational expression of T-bet and promotes Th1 cell responses mediating aplastic anemia in mice. J Immunol 192(11):5012–5022
Zhao E et al (2016) Cancer mediates effector T cell dysfunction by targeting microRNAs and EZH2 via glycolysis restriction. Nat Immunol 17(1):95–103
He S et al (2017) Ezh2 phosphorylation state determines its capacity to maintain CD8(+) T memory precursors for antitumor immunity. Nat Commun 8(1):2125
Gunawan M et al (2015) The methyltransferase Ezh2 controls cell adhesion and migration through direct methylation of the extranuclear regulatory protein talin. Nat Immunol 16(5):505–516
Shi X et al (2016) Epigenetic suppression of the antitumor cytotoxicity of NK cells by histone deacetylase inhibitor valproic acid. Am J Cancer Res 6(3):600–614
Santourlidis S et al (2002) Crucial role of DNA methylation in determination of clonally distributed killer cell Ig-like receptor expression patterns in NK cells. J Immunol 169(8):4253–4261
Santourlidis S et al (2008) Lineage-specific transition of histone signatures in the killer cell Ig-like receptor locus from hematopoietic progenitor to NK cells. J Immunol 180(1):418–425
Nagel S et al (2010) Polycomb repressor complex 2 regulates HOXA9 and HOXA10, activating ID2 in NK/T-cell lines. Mol Cancer 9:151
Yin J et al (2015) Ezh2 regulates differentiation and function of natural killer cells through histone methyltransferase activity. Proc Natl Acad Sci USA 112(52):15988–15993
DuPage M et al (2015) The chromatin-modifying enzyme Ezh2 is critical for the maintenance of regulatory T cell identity after activation. Immunity 42(2):227–238
Wang D et al (2018) Targeting EZH2 reprograms intratumoral regulatory T cells to enhance cancer immunity. Cell Rep 23(11):3262–3274
Acknowledgements
We thank Catherine Crowley-Kühn for critical reading of the manuscript. This work was funded by the Sassella Foundation (to N. A. R.), Swiss National Science Foundation Grant 310030-172978 (to O. B.), Swiss Cancer Research Grant KFS-4136-02-2017 (to O. B.), and Hochspezialisierte Medizin Schwerpunkt Immunologie (HSM-2-Immunologie; to O. B.).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Arenas-Ramirez, N., Sahin, D. & Boyman, O. Epigenetic mechanisms of tumor resistance to immunotherapy. Cell. Mol. Life Sci. 75, 4163–4176 (2018). https://doi.org/10.1007/s00018-018-2908-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-018-2908-7