Abstract
Aim and objective
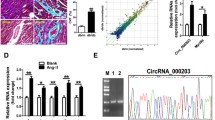
Regulation of microRNA gene expression by DNA methylation may represent a key mechanism to drive cardiac fibrosis progression. Cardiac fibroblast autophagy is the primary source of cardiac fibrosis, but the mechanisms underlying this process are incompletely understood. Here we found that DNMT3A suppression of the microRNA-200b (miR-200b) through pathway leads to cardiac fibroblast autophagy in cardiac fibrosis.
Methods
To understand the impact of DNMT3A on miR-200b at cardiac fibrosis, the rat cardiac fibrosis model was established via the abdominal aortic coarctation. Cardiac fibroblasts (CFs) were harvested from SD neonate rats and cultured. The expression of DNMT3A, miR-200b, collagen I was measured by western blotting, immunohistochemistry and qRT-PCR. Gain- or loss-of-function approaches were used to manipulate DNMT3A and miR-200b.
Results
DNMT3A level was upregulated and negatively correlated with miR-200b expression in fibrosis tissues and cardiac fibroblast. We found that autophagy was activated by miR-200b inhibitor and inactivated by miR-200b mimic in the rat cardiac fibroblast. Knockdown of DNMT3A notably increased the expression of miR-200b.
Conclusions
Taken together, these findings indicate that DNMT3A regulation of miR-200b controls cardiac fibroblast autophagy during cardiac fibrosis and provide a basis for the development of therapies for cardiac fibrosis.





Similar content being viewed by others
Abbreviations
- miR-200b:
-
MicroRNA-200b
- ECM:
-
Extracellular matrix
- CFs:
-
Cardiac fibroblasts
- DNMT3A:
-
DNA methyltransferases 3A
- H&E:
-
Hematoxylin-eosin
- siRNA:
-
Small interfering RNA
- AO:
-
Acridine orange
- PVDF:
-
Polyvinylidene fluoride
References
Molkentin JD, Bugg D, Ghearing N, Dorn LE, Kim P, Sargent MA, et al. Fibroblast-specific genetic manipulation of p38 mitogen-activated protein kinase in vivo reveals its central regulatory role in fibrosis. Circulation. 2017;136:549–61.
Christensen G, Herum KM, Lunde IG. Sweet, yet underappreciated: proteoglycans and extracellular matrix remodeling in heart disease. Matrix Biol. 2018.
Kaur H, Takefuji M, Ngai CY, Carvalho J, Bayer J, Wietelmann A, et al. Targeted ablation of periostin-expressing activated fibroblasts prevents adverse cardiac remodeling in mice. Circ Res. 2016;118:1906–17.
Peraro L, Zou Z, Makwana KM, Cummings AE, Ball HL, Yu H, et al. Diversity-oriented stapling yields intrinsically cell-penetrant inducers of autophagy. J Am Chem Soc. 2017;139:7792–802.
Yuan Y, Zhang Y, Han X, Li Y, Zhao X, Sheng L. Relaxin alleviates TGFbeta1-induced cardiac fibrosis via inhibition of Stat3-dependent autophagy. Biochem Biophys Res Commun. 2017;493:1601–7.
Yin S, Zhang Q, Yang J, Lin W, Li Y, Chen F, et al. TGFbeta-incurred epigenetic aberrations of miRNA and DNA methyltransferase suppress Klotho and potentiate renal fibrosis. Biochimica et biophysica acta. 2017;1864:1207–16.
Zou M, Wang F, Gao R, Wu J, Ou Y, Chen X, et al. Autophagy inhibition of hsa-miR-19a-3p/19b-3p by targeting TGF-beta R II during TGF-beta1-induced fibrogenesis in human cardiac fibroblasts. Sci Rep. 2016;6:24747.
Verjans R, Peters T, Beaumont FJ, van Leeuwen R, van Herwaarden T, Verhesen W, et al. MicroRNA-221/222 family counteracts myocardial fibrosis in pressure overload-induced heart failure. Hypertension. 2018;71:280–8.
Benati M, Montagnana M, Danese E, Paviati E, Giudici S, Franchi M, et al. Evaluation of mir-203 expression levels and DNA promoter methylation status in serum of patients with endometrial cancer. Clin Lab. 2017;63:1675–81.
Mayer SC, Gilsbach R, Preissl S, Monroy Ordonez EB, Schnick T, Beetz N, et al. Adrenergic repression of the epigenetic reader MeCP2 facilitates cardiac adaptation in chronic heart failure. Circ Res. 2015;117:622–33.
Ghosh AK, Rai R, Flevaris P, Vaughan DE. Epigenetics in reactive and reparative cardiac fibrogenesis: the promise of epigenetic therapy. J Cell Physiol. 2017;232:1941–56.
Cole CB, Russler-Germain DA, Ketkar S, Verdoni AM, Smith AM, Bangert CV, et al. Haploinsufficiency for DNA methyltransferase 3A predisposes hematopoietic cells to myeloid malignancies. J Clin Investig. 2017;127:3657–74.
Hertig V, Tardif K, Meus MA, Duquette N, Villeneuve L, Toussaint F, et al. Nestin expression is upregulated in the fibrotic rat heart and is localized in collagen-expressing mesenchymal cells and interstitial CD31(+)- cells. PloS One. 2017;12:e0176147.
Olivares-Silva F, Landaeta R, Aranguiz P, Bolivar S, Humeres C, Anfossi R, et al. Heparan sulfate potentiates leukocyte adhesion on cardiac fibroblast by enhancing Vcam-1 and Icam-1 expression. Biochimica et Biophysica Acta. 2017;1864:831–42.
Takemoto Y, Ramirez RJ, Kaur K, Salvador-Montanes O, Ponce-Balbuena D, Ramos-Mondragon R, et al. Eplerenone reduces atrial fibrillation burden without preventing atrial electrical remodeling. J Am Coll Cardiol. 2017;70:2893–905.
Cabrera S, Maciel M, Herrera I, Nava T, Vergara F, Gaxiola M, et al. Essential role for the ATG4B protease and autophagy in bleomycin-induced pulmonary fibrosis. Autophagy. 2015;11:670–84.
Chen TH, Chen MR, Chen TY, Wu TC, Liu SW, Hsu CH, et al. Cardiac fibrosis in mouse expressing DsRed tetramers involves chronic autophagy and proteasome degradation insufficiency. Oncotarget. 2016;7:54274–89.
Li XY, Wang SS, Han Z, Han F, Chang YP, Yang Y, et al. Triptolide restores autophagy to alleviate diabetic renal fibrosis through the miR-141-3p/PTEN/Akt/mTOR pathway. Mol Ther Nucl Acids. 2017;9:48–56.
Chen J, Zhang H, Hu J, Gu Y, Shen Z, Xu L, et al. Hydrogen-rich saline alleviates kidney fibrosis following AKI and retains Klotho expression. Front Pharmacol. 2017;8:499.
Acknowledgements
This work was supported by grants from National Nature Scientific Foundation of China (no. 81700212, no. 81570295) and Natural Science Foundation of Anhui Provincial Education Department (KJ2017A168) and Natural Science Foundation of Anhui Provincial (1808085MH231).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The author declares that there is no competing interest.
Additional information
Responsible Editor: John Di Battista.
Rights and permissions
About this article
Cite this article
Zhao, XD., Qin, RH., Yang, JJ. et al. DNMT3A controls miR-200b in cardiac fibroblast autophagy and cardiac fibrosis. Inflamm. Res. 67, 681–690 (2018). https://doi.org/10.1007/s00011-018-1159-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00011-018-1159-2