Abstract
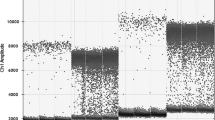
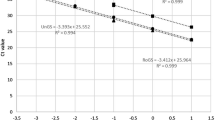
The detection of genetically modified organisms in feedstuffs using PCR methods rely on the yield and quality of the extracted genomic plant DNA. The used extraction methods must be able to provide DNA of sufficient amounts of non-fragmented DNA that is free of PCR inhibitors. This article describes the comparison of methods for the extraction of DNA from maize gluten as an example for a protein-rich corn-based feedstuff sample and the following validation of two selected methods. These selected methods are based on lysis by a cetyltrimethylammonium bromide buffer followed by precipitation of genomic DNA and on a kit-based procedure. To calculate the amount of amplifiable DNA copies and the PCR efficiency in the extracts, the crossing points of the real-time PCR curves were used. The comparison of different extraction methods showed that the two selected methods yielded the highest amounts of amplifiable DNA based on the amplification of the maize specific reference gene hmg by real-time TaqMan PCR. Additionally, approaches to increase DNA yield or concentration by variation of the protocol are discussed. Both validation studies were conducted by the working group “PCR analytics in feed” of the Association of German Agricultural and Research Institutes.


Similar content being viewed by others
References
Abdel-Latif A, Osman G (2017) Comparison of three genomic DNA extraction methods to obtain high DNA quality from maize. Plant Methods 3:1. https://doi.org/10.1186/s13007-016-0152-4
Broeders S, Huber I, Grohmann L, Berben G, Taverniers I, Mazzara M, Roosens N, Morriset D (2014) Guidelines for validation of qualitative real-time PCR methods. Trends Food Sci Technol 37:115–116. https://doi.org/10.1016/j.tifs.2014.03.008
Delobel C, Larcher S, Mazzara M, Van Den Eede G (2008) Event-specific method for the quantification of maize event Bt11 using real-time PCR—Validation Report And Protocol. https://doi.org/10.2788/4370
Demeke T, Jenkins GR (2009) Influence of DNA extraction methods, PCR inhibitors and quantification methods on real-time PCR assay of biotechnology-derived traits. Anal Bioanal Chem 396(6):1977–1990. https://doi.org/10.1007/s00216-009-3150-9
Domey S, Eichner C, Speck B (2018) Konzept zur Analytik von gentechnisch veränderten Futtermitteln—Arbeitspapier des Arbeitskreises PCR-Analytik der Fachgruppe VI des Verbandes Deutscher Landwirtschaftlicher Untersuchungs- und Forschungsanstalten (VDLUFA); Version 3, Stand Dezember 2018
Graziolo E, Bonfini L, Pinski G, Van den Ede G (2009) Report on the Validation of a DNA-Extraction Method for Soybean Seeds. Joint Research Centre, European Commission, CRLVL04/07XP
Gryson N (2010) Effect of food processing on plant DNA degradation and PCR-based GMO analysis: a review. Anal Bioanal Chem 396(6):2003–3022. https://doi.org/10.1007/s00216-009-3343-2
ISAAA (2018) Global Status of Commercialized Biotech/GM Crops: 2018. ISAAA Brief No. 54. ISAAA: Ithaca, NY
ISO 21571:2013–08 Foodstuffs—Methods of analysis for the detection of genetically modified organisms and derived products—Nucleic acid extraction. ISO 21571:2005 + Annex1:2013
JRC–Joint Research Centre, Institute for Health (2011) Verification of analytical methods for GMO testing when implementing interlaboratory validated methods—Guidance document from the European Network of GMO laboratories (ENGL), prepared by the ENGL working group on “Method Verification”. EUR 24790 EN–2011
JRC—Joint Research Centre, European Commission, European Network of GMO Laboratories (ENGL) (2015) Definition of Performance Requirements for Analytical Methods of GMO Testing. JRC Technical Report
Lottspeich F, Engels JW (2012) Bioanalytik, 3. Spektrum Akademischer Verlag Heidelberg, Auflage
Mazzara M, Graziolo E, Savini C, Van Den Eede G (2009): Report on the Verification of the Performance of a MON810 Event-specific Method on Maize Line MON810 Using Real-time PCR - Validation Report and Protocol. https://doi.org/10.2788/59036
Official collection of test methods (2010) Method for the Detection of nucleic acid sequences using Polymerase Chain Reaction (PCR), General information and requirements. German law on genetic engineering, Article 28b, G 00.00–5
Pacheco Coello R, Pestana Justo J, Factos Mendoza A, Santos Ordonaz E (2017) Comparison of three DNA extraction methods for the detection and quantification of GMO in Ecuadorian manufactured food. BMC Res Notes 10(1):789. https://doi.org/10.1186/s13104-017-3083-x
Pfaffl MW (2004) Real-time RT-PCR: Neue Ansätze zur exakten mRNA Quantifizierung. BIOspektrum Sonderausgabe PCR 10(04):92–95
Positive List for Straight Feeding Stuffs 13th Ed. (2019) Standards Commission for Straight Feeding Stuffs at the Central Committee of the German Agriculture
Promega Corporation (2010) Wizard DNA Clean-up System. Instructions for use of product A7280. Part #TB141. Promega Corporation, www.promega.com
Rasmussen R (2001) Quantification on the LightCycler. In: Meuer S, Wittwer C, Nakagawara (Eds) Rapid Cycle Real-Time PCR, Methods and Applications. 1st edn. Springer, Berlin, pp 21–34
Savini C, Mazzara M, Pinski G, Van den Eede, G (2012) Event-specific Method for the Quantification of Maize MON 87460 Using Real-time PCR—Validation Report and Protocol. https://doi.org/10.2788/45380
Tung Nguyen CT, Son R, Raha AR, Lai OM, Clemente Michael WVL (2009) Comparison of DNA extraction efficiencies using various methods for the detection of genetically modified organisms (GMOs). Int Food Res J 16:21–30
Turkec A, Kazan H, Karancanli B, Lucas SJ (2015) DNA extraction techniques compared for accurate detection of genetically modified organisms (GMOs) in maize food and feed products. J Food Sci Technol 52(8):5164–5171. https://doi.org/10.1007/s13197-014-1547-8
VDLUFA (2012) Detection of animal components in feed using molecular biology methods. Method Collect Part III Feedstuffs Method 29:1
Waiblinger HU, Schillinger M, Hess N (2012) In-house and interlaboratory validation of a method for the extraction of DNA from maize starch. J Verbr Lebensm 7(2):163–170. https://doi.org/10.1007/s00003-012-0765-0
Acknowledgements
We want to thank all laboratories that participated in the ring trials: Hessian State Laboratory (Kassel, Germany), Thuringian State Institute for Agriculture (Jena, Germany), Wessling GmbH (Altenberge, Germany), Austrian Agency for Health and Food Safety, Institute for Food Safety (Wien, Austria), National Reference Laboratory for Animal Protein in Feed, German Federal Institute for Risk Assessment (Berlin, Germany), National Reference Laboratory for Genetically Modified Organisms, Federal Office of Consumer Protection and Food Safety (Berlin, Germany), Bavarian Health and Food Safety Authority (Oberschleissheim, Germany), Chemical and Veterinary Investigatory Office Rhein-Ruhr-Wupper (Krefeld, Germany), Chemical and Veterinary Investigatory Office OWL (Detmold, Germany), Institute for Hygiene and Environment (Hamburg, Germany), Saarland State Office for Consumer Protection (Saarbrücken, Germany). LUFA Speyer (Speyer, Germany), Biolytix AG (Witterswil, Switzerland). In addition we want to thank Benjamin Pickel (LUFA Speyer, Speyer, Germany), Ole Sindt (State Laboratory Schleswig Holstein, Neumuenster, Germany), and Ulf-Martin Kohlstock (State Agency for Agriculture and Horticulture Sachsen-Anhalt) for critically reading the manuscript. The Federal Office of Consumer Protection and Food Safety (Berlin, Germany) provided the chemicals required for the realization of the ring trials.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Matthes, N., Westphal, K., Haldemann, C. et al. Validation of a modified CTAB method for DNA extraction from protein-rich maize feedstuffs. J Consum Prot Food Saf 15, 331–340 (2020). https://doi.org/10.1007/s00003-020-01285-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00003-020-01285-y