Abstract
Background and Objectives: Drug resistance in HIV-1 is one of the main causes of failure of antiretroviral therapy. Phenotypic detection of drug-resistant HIV-1 can provide guidance in selecting the optimal treatment regimen. Traditional phenotype assays are labor intensive and time consuming. Thus, a rapid and convenient phenotype assay with a single cycle of replication was developed and used in this study.
Methods: Two restriction endonuclease sites, ApaI and AgeI, were inserted into the plasmid pSG3,DΔenv using site-directed mutagenesis. The reverse transcriptase and protease genes of HIV-1 were amplified from patients and cloned into the modified pSG3Δenv. Sixteen original recombinant pseudoviruses were generated. The phenotypic susceptibility of these 16 recombinant pseudoviruses to 12 antiretro viral drugs was determined using a luciferase reporter system, and the phenotype and genotype results were compared.
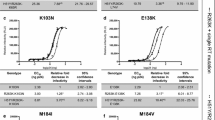
Results: A modified phenotype assay with a single-cycle system was established, and its reproducibility and feasibility were validated. Approximately 89% of the phenotype results were in agreement with the genotype results; this slight disagreement may have been due to complex and multiple resistance mutations. The phenotype results showed that individual pseudoviruses with four thymidine analog mutations (TAMs) [M41L, T67N, L210W, and T215Y] in combination with various other mutations had different levels of resistance to nucleoside reverse transcriptase inhibitors (NRTIs). Mutations E44A, T69D, and V118I influenced the pattern of resistance of TAMs. The level of resistance to non-NRTIs (NNRTIs) was also variable when different NNRTI-resistance mutations were combined.
Conclusion: The single-cycle pseudovirus phenotypic susceptibility detection system reflects HIV-1 drug resistance, especially for complex resistance mutants, and could be used to screen new antiretroviral candidates.




Similar content being viewed by others
References
Mohammadpour A, Yekta ZP, Nikbakht Nasrabadi AR. HIV-infected patients' adherence to highly active antiretroviral therapy: a phenomeno-logical study. Nurs Health Sci 2010; 12: 464–9
Grover D, Copas A, Green H, et al. What is the risk of mortality following diagnosis of multidrug-resistant HIV-1? J Antimicrob Chemother 2008; 61: 705–13
Gallant JE. Antiretroviral drug resistance and resistance testing. Top HIV Med 2005; 13: 138–42
Hertogs K, de Bethune MP, Miller V, et al. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob Agents Chemother 1998; 42: 269–76
Petropoulos CJ, Parkin NT, Limoli KL, et al. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob Agents Chemother 2000; 44: 920–8
Zhang H, Zhou Y, Alcock C, et al. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. J Virol 2004; 78: 1718–29
McMahon MA, Shen L, Siliciano RF. New approaches for quantitating the inhibition of HIV-1 replication by antiviral drugs in vitro and in vivo. Curr Opin Infect Dis 2009; 22: 574–82
Covens K, Dekeersmaeker N, Schrooten Y, et al. Novel recombinant virus assay for measuring susceptibility of human immunodeficiency virus type 1 group M subtypes to clinically approved drugs. J Clin Microbiol 2009; 47: 2232–42
Kutner RH, Zhang XY, Reiser J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat Protoc 2009; 4: 495–505
Garcia-Perez J, Sanchez-Palomino S, Perez-Olmeda M, et al. A new strategy based on recombinant viruses as a tool for assessing drug susceptibility of human immunodeficiency virus type 1. J Med Virol 2007; 79: 127–37
Derdeyn CA, Decker JM, Sfakianos JN, et al. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J Virol 2000; 74: 8358–67
Takeuchi Y, McClure MO, Pizzato M. Identification of gammaretroviruses constitutively released from cell lines used for human immunodeficiency virus research. J Virol 2008; 82: 12585–8
Wei X, Decker JM, Liu H, et al. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother 2002; 46: 1896–905
Wei X, Decker JM, Wang S, et al. Antibody neutralization and escape by HIV-1. Nature 2003; 422: 307–12
Chong H, Hong K, Zhang C, et al. Genetic and neutralization properties of HIV-1 env clones from subtype B/BC/AE infections in China. J Acquir Immune Defic Syndr 2008; 47: 535–43
Betts BJ, Shafer RW. Algorithm specification interface for human immunodeficiency virus type 1 genotypic interpretation. J Clin Microbiol 2003; 41: 2792–4
Kantor R, Machekano R, Gonzales MJ, et al. Human immunodeficiency virus reverse transcriptase and protease sequence database: an expanded data model integrating natural language text and sequence analysis programs. Nucleic Acids Res 2001; 29: 296–9
Koup RA, Ho DD, Poli G, et al. Isolation and quantitation of HIV in peripheral blood. Curr Protoc Immunol 2001; Chapter 12: Unit 12.2
Rhee SY, Taylor J, Wadhera G, et al. Genotypic predictors of human immunodeficiency virus type 1 drug resistance. Proc Natl Acad Sci U S A 2006; 103: 17355–60
Ceccherini-Silberstein F, Svicher V, Sing T, et al. Characterization and structural analysis of novel mutations in human immunodeficiency virus type 1 reverse transcriptase involved in the regulation of resistance to nonnucleoside inhibitors. J Virol 2007; 81(20): 11507–19
Girouard M, Diallo K, Marchand B, et al. Mutations E44D and V118I in the reverse transcriptase of HIV-1 play distinct mechanistic roles in dual resistance to AZT and 3TC. J Biol Chem 2003; 278: 34403–10
Gianotti N, Galli L, Boeri E, et al. The 118I reverse transcriptase mutation is the only independent genotypic predictor of virologic failure to a stavudine-containing salvage therapy in HIV-1-infected patients. J Acquir Immune Defic Syndr 2006; 41: 447–52
Deshpande A, Jauvin V, Magnin N, et al. Resistance mutations in subtype C HIV type 1 isolates from Indian patients of Mumbai receiving NRTIs plus NNRTIs and experiencing a treatment failure: resistance to AR. AIDS Res Hum Retroviruses 2007; 23: 335–40
Ferradini L, Jeannin A, Pinoges L, et al. Scaling up of highly active antiretroviral therapy in a rural district of Malawi: an effectiveness assessment. Lancet 2006; 367: 1335–42
Jackson JB, Becker-Pergola G, Guay LA, et al. Identification of the K103N resistance mutation in Ugandan women receiving nevirapine to prevent HIV-1 vertical transmission. AIDS 2000; 14: F111–5
Jourdain G, Ngo-Giang-Huong N, Le CS, et al. Intrapartum exposure to nevirapine and subsequent maternal responses to nevirapine-based antiretroviral therapy. N Engl J Med 2004; 351: 229–40
Kantor R, Katzenstein DA, Efron B, et al. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: results of a global collaboration. PLoS Med 2005; 2: e1 12
Kassaye S, Lee E, Kantor R, et al. Drug resistance in plasma and breast milk after single-dose nevirapine in subtype C HIV type 1: population and clonal sequence analysis. AIDS Res Hum Retroviruses 2007; 23: 1055–61
Grossman Z, Istomin V, Averbuch D, et al. Genetic variation at NNRTI resistance-associated positions in patients infected with HIV-1 subtype C. AIDS 2004; 18: 909–15
Whitcomb JM, Parkin NT, Chappey C, et al. Broad nucleoside reversetranscriptase inhibitor cross-resistance in human immunodeficiency virus type 1 clinical isolates. J Infect Dis 2003; 188: 992–1000
Acknowledgments
The authors would like to thank Mr Zuoyi Bao and Dr Jue Li from the Institute of Microbiology and Epidemiology at the Academy of Military Medical Science (Beijing, P.R. China) and Mr Yile Xue from the Shanghai Center of Disease Control (Shanghai, P.R. China) for their technical assistance, and Dr Fujie Zhang from the National Center of Disease Control (Beijing, P.R. China) for his suggestions.
This study was supported by the Key Project on Infectious Diseases such as AIDS, Hepatitis, and Tuberculosis (grant no. 2009ZX10004-801) from the Ministry of Science and Technology (Beijing, P.R. China). The authors declare no conflicts of interest that are directly relevant to the content of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jia, Z., Xu, S., Nie, J. et al. Phenotypic Analysis of HIV-1 Genotypic Drug-Resistant Isolates from China, Using a Single-Cycle System. Mol Diag Ther 15, 293–301 (2011). https://doi.org/10.1007/BF03256421
Published:
Issue Date:
DOI: https://doi.org/10.1007/BF03256421