Abstract
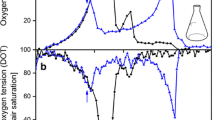
The response of IPTG induction was investigated through the monitoring of the alkali consumption rate and buffer capacity during the cultivation of recombinantE. coli BL21(DE3) harboring the plasmid pRSET-LacZ under the control oflac promoter. The rate of alkali consumption increased along with cell growth, but declined suddenly after approximately 0.2 h of IPTG induction. The buffer capacity also declined after 0.9 h of IPTG induction. The profile of buffer capacity seems to correlate with the level of acetate production. The IPTG response was monitored only when introduced into the mid-exponential phase of bacterial cell growth. The minimum concentration of IPTG for induction, which was found out to be 0.1 mM, can also be monitored on-line andin-situ. Therefore, the on-line monitoring of alkali consumption rate and buffer capacity can be an indicator of the metabolic shift initiated by IPTG supplement, as well as for the physiological state of cell growth.
Similar content being viewed by others
References
Schügerl, K. (1991) Common instruments for process analysis and control. pp. 6–25, In: H. J. Rehm, G. Reed, A. Puhler, and P. Stadler (eds.)Biotechnology 4. VCH Publishers Inc., NY, USA.
Suzuki, T., T. Yamane, and S. Shimizu (1990) Phenomenological background and some preliminary trials of automated substrate supply in pH-stat model fed-batch culture using a set point of high limit.J. Ferment. Bioeng. 69: 292–297.
Chung, Y. and W. Hur (2000) A new method of measuring of buffer capacity and alkali consumption rate of a fermentation process.J. Bioscience. Bioeng. 90: 580–582.
San, K. and G. Stephanopoulos (1984) Studies on on-line bioreactor identification IV. Utilization of pH measurement for product estimation.Biotechnol. Bioeng. 26: 1209–1218.
Antonio, V., I. C. Juan, A. T. Jose, and U. Unai (1998) On-line estimation of biomass through pH control analysis in aerobic yeast fermentation systems.Biotechnol. Bioeng. 58: 445–450.
Hur, W. (1997) Mathematical analysis on the pH change during cell growth in a phosphate buffer based medium, Korean.J. Biotechnol. Bioeng. 12: 167–175.
Han, K. (1992)A Study of Acetic Acid Formation in Escherichia coliFermentation. Ph. D. Thesis. University of California, Irvine, CA, USA.
Luli, G. W. and W. R. Strohl (1990) Comparison of growth, acetate production, and acetate inhibition ofEscherichia coli strains in batch and fed-batch fermentations.Appl. Environ. Microbiol. 56: 1004–1011.
Sun, W.-J., C. Lee, H. A. George, A. L. Powell, M. E. Dahlgren, R. Gresham, and C. H. Park (1993) Acetate inhibition on growth of recombinantE. coli and expression of fusion protein TGFa-PE40.Biotechnol. Lett. 15: 809–814.
Miller, G. L. (1959) Use of dinitrosalicylic acid reagent for determination of reduction sugar.Anal. Chem. 31: 426–428.
Studier, F. W. and B. A. Moffatt (1986) Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes.J. Mol. Biol. 189: 113–130.
Glick, B. R. (1995) Metabolic load and heterologous gene expression.Biotechnol. Adv. 13: 247–261.
Zaslaver, A., A. E. Mayo, R. Rosenberg, P. Bashkin, H. Sberro, M. Tsalyuk, M. G. Surette, and U. Alon (2004) Just-in-time transcription program in metabolic pathways.Nature Genetics 36: 486–491.
Kosinski, M. J., U. Rinas, and J. E. Bailey (1992) Isopropyl-β-d-thiogalactopyranoside influences the metabolism ofEscherichia coli.Appl. Environ. Microbiol. 36: 782–784.
Xu, Z., G. Liu, P. Cen, and W. K. R. Wong (2000) Factors influencing excretive production of human epidermal growth factor (hEGF) with recombinantEscherichia coli K12 system.Bioprocess. Biosystem Eng. 23: 669–674.
Lee, C., W.-J. Sun, B. W. Burgess, B. H. Junker, J. Reddy, B. C. Buckland, and R. L. Greasham (1997) Process optimization for large scale production of TGF-α-PE40 in recombinantEscherichia coli: Effect of medium composition and induction timing on protein expression.J. Ind. Microbiol. Biotechnol. 18: 260–266.
Sivakesava, S., Z. N. Xu, Y. H. Chen, J. Hackett, R. C. Huang, E. Lam, T. L. Lam, K. L. Siu, R. S. C. Wong, and W. K. R. Wong (1999) Production of excreted human epidermal growth factor (hEGF) by an efficient recombinantEscherichia coli system.Process Biochem. 34: 893–900.
Donovan, R. S., C. W. Robinson, and B. R. Glick (1996) Review: Optimizing inducer and culture conditions for expression of foreign proteins under the control of the lac promoter.J. Ind. Microbiol. 16: 145–54.
Studier, F. and B. Moffatt (1986) Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes.J. Mol. Biol. 189: 113–130.
Xie, L., D. Hall, M. A. Etieman, and E. Altman (2003) Optimization of recombinant aminolevulinate synthase production inEscherichia coli using factorial design.Appl. Microbial. Biotechnol. 63: 267–273.
Kosinski, M. J., U. Rinas, and J. E. Bailey (1992) Isopropyl-β-d-thiogalactopyranoside influences the metabolism ofEscherichia coli.Appl. Microbiol. Biotechnol. 36: 782–783.
Donovan, R. S., C. W. Robinson, and B. R. Glick (2000) Optimizing the expression of a monoclonal antibody fragment under the transcriptional control of theEscherichia coli lac promoter.Can. J. Microbiol. 46: 532–541.
Ramisetti, S., H. A. Kang, S. K. Lee, and C. H. Kim (2003) Production of recombinant hirudin in galactokinase-deficientSaccharomyces cerevisiae by fed-batch fermentation with continuous glucose feeding.Biotechnol. Bioprocess Eng. 8: 183–186.
Kim, C. H., J. Rao, D. J. Youn, and S. K. Lee (2003) Scale-up of recombinant hirudin production fromSaccharomyces cerevisiae.Biotechnol. Bioprocess Eng. 8: 303–305.
Albano, C. R., L. Randers-Eichhorn, W. E. Bentley, and G. Rao (1998) Green fluorescent protein as a real time quantitative reporter of heterologous protein production.Biotechnol. Prog. 14: 350–354.
Jones, J. J., A. M. Bridges, A. P. Fosberry, S. Gardner, R. R. Lowers, R. R. Newby, P. J. James, R. M. Hall, and O. Jenkins (2004) Potential of real-time measurement of GFP-fusion proteins.J. Biotechnol. 109: 201–211.
Rhee, J. I., A. Ritzka, and T. Scheper (2004) On-line monitoring and control of substrate concentrations in biological processes by flow injection analysis systems.Biotechnol. Bioprocess Eng. 9: 156–165.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hur, W., Chung, YK. On-line monitoring of IPTG induction for recombinant protein production using an automatic pH control signal. Biotechnol. Bioprocess Eng. 10, 304–308 (2005). https://doi.org/10.1007/BF02931846
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF02931846