Summary
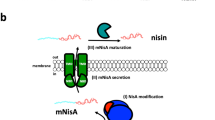
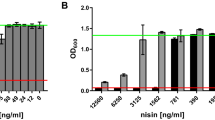
In this study, a plasmid was integrated into nisP, creating the first defined mutation in a nisin biosynthetic gene. The mutant strain secreted fully modified nisin with the N-terminal leader still attached. The presence of the leader was corfirmed by N-terminal sequencing of the purified precursor. The dehydration and lanthiomne formation of the precursor were already completed as active nisin could be formed by cleaving the leader from the inactive precursor by a trypsin treatment or by incubation with wild type cells. Nisin immunity of the NisP mutant strain was lowered to about 10% of the wild type immunity. The results show that NisP is needed for precursor processing and for development of high immunity of nisin.
Similar content being viewed by others
References
Buchman W B, Banerjee S, Hansen J N. Structure, expression and evolution of a gene encoding the precursor of nisin, a small protein antibiotic. J Biol Chem, 1988, 263: 16 260
Dodd H M, Horn N, Gasson M J. Analysis of the genetic determinant for production of the peptide antibiotic nisin. J Gen Microbiol, 1990, 136: 555
Klein C, Kaletta C, Entian K -D. Biosynthesis of the lantibiotic subtilin is regulated by a histidine kinase/response regulator system. Appl Environm Microbiol, 1993, 59: 296
Van der Meer J, Polman J, Beerthuyen M Met al. Characterization of the Lactococcus lactis nisin A operon genes nisP, encoding a subtilisin-like serine protease involved in precursor processing, and nisR, encoding a regulatory protein involved in nisin biosynthesis. J Bacteriol, 1993, 175: 2578
Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their abcteriophages. Appl Microbiol, 1975, 29: 807
Marmur J. A procedure for the isolation of deoxyribonucleic acids from micro-organisms. J Mol Biol, 1961, 3: 208
Schägger H, von Jagow G. Tricinesodium dodccyl sulphatepolyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biocheml, 1987, 166: 368
Yang, R, Johnson M C, Ray B. Novel method to extractlarge amouns of bacteriocins from lactic acid bacteria. Appl Environm Microbiology, 1992, 58: 3355
Mulders J W M, Boerrighter I J, Rollema H Set al. Identification and characterization of the lantibiotic nisin Z, a natural nisin variant. Eur J Biochem, 1991, 201: 591
Genetics Computer Group. Program manual for the GCG package. Version 7. 575 Science Drive, Madison, Wisconsin, April 1991
Author information
Authors and Affiliations
Additional information
This work was supported by the University of Helsinki and Valio Ltd and conducted in Finland
Rights and permissions
About this article
Cite this article
Si-ying, Y., Koponen, O., Qiao, M. et al. NisP is related to nisin precursor processing and possibly to immunity in lactococcus lactis. Journal of Tongji Medical University 15, 193–197 (1995). https://doi.org/10.1007/BF02887942
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF02887942