Abstract
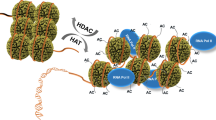
In eukaryotes, nucleosome is the basic unit of chromatin. Nucleosorne is composed of an octamer of histone proteins (two molecules each of histones H2A, H2B, H3 and H4) and DNA strand wound around the octamer. Some data show that core histone octamer can affect gene transcription bothin vitro andin vivo. Recent results indicate that histone acetylation/deacetylation is a key step to regulate activity of genes. This article summarizes some coactivators, such as GCNSp, P300/CBP and TAFII 250, which are recently found to have histone acetyltransferase activity. The relationship between these coactivators and gene activation is also described. Besides, this article concerns some corepressors which have histone deacetylase activity, such as Rpd3p, HDAC2. These corepressors combine with other protein complex and then repress transcription. Finally, some problems to be solved and the future direction in this active field are discussed.
Similar content being viewed by others
References
Workman, J. L., Roeder, R. G., Binding of transcription factor TFII D to the major late promoter duringin vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II,Cell, 1987, 51: 613.
Meisteremst, M., Horikoshi, M., Roeder, R. G., Recombinant yeast TFIID, a general transcription factor, mediates activation by the gene-specific factor USF in a chromatin assembly assay,Proc. Natl. Acad. Sci. USA, 1990, 87: 9135.
Godde, J. S., Nakatani, Y., Wolffe, A. P., The amino-terminal tails of the core histories and the translational position of the TATA box determine TBP/TFIIA association with nucleosomal DNA,Nucleic Acids Res., 1995, 23: 4557.
Turner, B. M., Girley, A. J., Lavender, J., Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei.Cell, 1992, 69; 375.
Hebbes, T. R., Clayton, A. I., Thome, A. W.et al., Core histone hyperacetylation co-maps with generalized DNasel sensitivity in the chicken ß-globin chromosomal domain,EMBO J., 1994, 13: 1823.
Braunstein, M., Rose, A. V., Holmes, S. G.et al., Transcriptional silencing in yeast is associated with reduced nucleosome acetylation,Genes Dev., 1993, 7: 592.
Garcia-Ramirez, M., Rocchine, C., Ausio, J., Modulation of chromatin folding by histone acetylation,J. Biol. Chem., 1995, 270: 17923.
Macus, G. A., Silverman, N., Berger, S. L.et al., Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors,EMBO J., 1994, 13: 4807.
Brownell, J. E., Zhou, J., Ranalli, T.et al., Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation.Cell, 1996, 84: 843.
Yang, X. J., Ogryzko, V. V., Nishikawa, J.et al., A p300/CBP-associated factor that competes with the adenoiral oncoprotein E1A,Nature, 1996, 382: 319.
Ogryazko, V. V., Schlitz, R. L., Russanova, V.et al., The transcriptional coactivators p300 and CBP are histone acetyltransferases,Cell, 1996, 87: 953.
Bannister, A. J., Kouzarides, T., The CBP co-activator is a histone acetyltransferase,Nature, 1996, 384: 641.
Janknecht, R., Hunter, T., A growing coactivator network,Nature, 1996, 383: 22.
Mizzen, C. A., Yang, X. J., Kokubo, T.et al., The TAFII 250 subunit of TFIID has histone acetyltransferase activity,Cell, 1996, 87: 1261.
Ptashne, M., Gann, A., Transcriptional activation by recruitment,Nature, 1997, 386: 569.
Barberis, A., Pearlberg, J., Simkovich, N.et al., Contact with a component of the polymerase II holoenzyme suffices for gene activation.Cell, 1995, 81: 359.
Gaudreau, L., Schmid, A., Blaschke, D.et al., RNA polymerase II holoenzyme recruitment is sufficient to remodel chromatin at the yeast PHO5 promoter,Cell, 1997, 89: 55.
Roth, S. Y., Allis, C. D., The subunit-exchange model of histone acetylation,Trends Cell Biol., 1996, 6: 371.
Segil, N., Guermah, M., Hoffmann, A.et al., Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization,Gene Dev., 1996, 10: 2389.
Vidal, M., Faber, R. F., Rpd3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyes cerevisiae,Mol. Cell. Biol., 1991, 11: 6317.
Stillman, D. J., Dorland, S., Yu, Y., Epistatic analysis of suppressor mutations that allow HO expression in the absence of the yeast SWI5 transcriptional activator,Genetics, 1994, 136: 781.
Taunton, J., Hassig, C. A., Schreiber, S. L., A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p,Science, 1996, 272: 408.
Yang, W. M., Inouye, C., Zeng, Y.et al., Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3,Proc. Natl. Acad. Sci. USA, 1996, 93: 12845.
Wane, H., Clark, I., Nicholson, P. R.et al., The S. Cerevisiae SIN3 gene, a negative regulator of HO, contains four paired amphipathic helical motifs.Mol. Cell. Biol., 1990, 10: 5927.
Kadosh, D., Struhl, K., Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 hietone deacetylase to target promoters,Cell. 1997, 89: 365.
Ayer, D. E., Kretmer, L., Eisenman, R. N., Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sim3,Cell. 1995, 80: 767.
Schreiber-Agus, N., Chin, L., Chen, K.et al., An mineterminal domain of Mxil mediites anti-Myc onmgenic activity and interacts with a homolcg of the yeast transcriptional repressor SIN3,Cell, 1995, 80: 777.
Heiel, T., Lavinsky, R. M., Mullen, T. M.et al., A complex containing N-CoR, did and histone deacetylase mediates transcriptional repression,Nature, 1997, 387: 43.
Alland, L., Muhle, R., Jr, H. H.et al., Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression.Nature. 1997. 387: 49.
Ha, C. A., Fleischer, T. C., Billin, A. W.et al., Histone deacetylase activity is required for full transcriptional repression by mSidA,Cell. 1997, 89: 341.
Laherty, C. D., Yang, W. M., Sun, J. M.et al., Histone deacetylases associated with the mSin3 corepressor mediite Mad transcriptional repression,Cell. 1997, 89: 349.
Zhang, Y., Iratni, R., Erdjument-Bromage, H.et al., Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex,Cell. 1997, 89: 357.
Nagy, L., Kao, H. Y., Chakravarti, D.et al., Nuclear receptor repression mediated by a complex containing SMRT. mSin3A. and hietone deacetylase.Cell, 1997, 89: 373.
De Rubatis, F., Kadosh, D., Henchoz, S., The hiitone deacetyh RPD3 counteracts genomic silencing in Drosophila and yeast,Nature, 1996. 384: 589.
Gu, W., Shi, X. L., Roeder, R. G., Synergistic activation of transcription by W and p53,Nature, 1997, 387: 819.
Lill, N. L., Grossman, S. R., Ginsberg, D.et al., Binding and modulation of p53 by P300/CBP coactivators,Nature, 1997, 387: 823.
Author information
Authors and Affiliations
About this article
Cite this article
Lu, Z., Wang, Y. Relationship between histone acetylation/deacetylation and gene transcription. Chin.Sci.Bull. 43, 1057–1063 (1998). https://doi.org/10.1007/BF02883072
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF02883072