Abstract
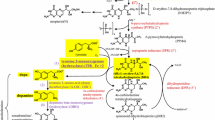
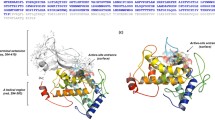
The neurotransmitter biosynthetic enzymes, tyrosine hydroxylase (TH), and tryptophan hydroxylase (TPH) are each composed of an amino-terminal regulatory domain and a carboxylterminal catalytic domain. A chimeric hydroxylase was generated by coupling the regulatory domain of TH (TH-R) to the catalytic domain of TPH (TPH-C) and expressing the recombinant enzyme in bacteria. The chimeric junction was created at proline 165 in TH and proline 106 in TPH because this residue is within a conserved five amino-acid span (ValProTrpPhePro) that defines the beginning of the highly homologous catalytic domains of TH and TPH. Radioenzymatic activity assays demonstrated that the TH-R/TPH-C chimera hydroxylates tryptophan, but not tyrosine. Therefore, the regulatory domain does not confer substrate specificity. Although the TH-R/TPH-C enzyme did serve as a substrate for protein kinase (PKA), activation was not observed following phosphorylation. Phosphorylation studies in combination with kinetic data provided evidence that TH-R does not exert a dominant influence on TPH-C. Stability assays revealed that, whereas TH exhibited a t1/2 of 84 min at 37°C, TPH was much less stable (t 1/2=28.3 min). The stability profile of TH-R/TPH-C, however, was superimposable on that of TH. Removal of the regulatory domain (a deletion of 165 amino acids from the N-terminus) of TH rendered the catalytic domain highly unstable, as demonstrated by at 1/2 of 14 min. The authors conclude that the regulatory domain of TH functions as a stabilizer of enzyme activity. As a corollary, the well-characterized instability of TPH may be attributed to the inability of its regulatory domain to stabilize the catalytic domain.
Similar content being viewed by others
References
Abate C. and Joh T. H. (1991) Limited proteolysis of rat brain tyrosine hydroxylase defines an N-terminal region required for regulation of cofactor binding and directing substrate specificity.J. Mol. Neurosci. 2, 203–215.
Abate C., Smith, J. A., and Joh T. H. (1988) Characterization of the catalytic domain of bovine adrenal tyrosine hydroxylase.Biochem. Biophys. Res. Commun. 151, 1446–1453.
Ausubel F. M., Brent R., Kingston R. E., Moore D., Seidman J. G., Smith J. A., and Struhl K. (1987)Current Protocols in Molecular Biology, Wiley, New York.
Beevers S. J., Knowles R. G., and Pogson C. I. (1983) A sensitive radiometric assay for tryptophan hydroxylase applicable to crude extracts.J. Neurochem. 40, 894–897.
Bonnefoy E., Ferrara P., Rohrer H., Gros F., and Thibault J. (1988) Role of the N-terminus of rat pheochromocytoma tyrosine hydroxylase in the regulation of the enzyme's activity.Eur. J. Biochem. 174, 685–690.
Cash C. D. P., Vayer P., Mandel P., and Maitre M. (1985) Tryptophan 5-hydroxylase: rapid purification from whole brain and production of specific antiserum.Eur. J. Biochem. 149, 239–245.
Cotton R. G. H., McAdam W., Jennings I., and Morgan F. J. (1988) A monoclonal antibody to aromatic amino acid hydroxylases: identification of the epitope.Biochem. J. 255, 193–196.
Daubner S. C., Lohse D. L., and Fitzpatrick P. F. (1993) Expression and characterization of catalytic and regulatory domains of rat tyrosine hydroxylase.Protein Sci. 2, 1452–1460.
Daubner S. C. and Fitzpatrick P. F. (1993) Lysine 241 of tyrosine hydroxylase is not required for binding of tetrahydrobiopterin substrate.Arch. Biochem. Biophys. 302, 455–460.
Daubner S. C. and Piper M. M. (1995) Deletion mutants of tyrosine hydroxylase identify a region critical for heparin binding.Protein Sci. 4, 538–541.
D'Sa C. M., Arthur R. E. Jr., and Kuhn D. M. (1996) Expression and deletion mutagenesis of tryptophan hydroxylase fusion proteins: delineation of the enzyme catalytic core.J. Neurochem. 67, 917–926.
Edelman A. M., Raese J. D., Lazar M. A., and Barchas J. D. (1981) Tyrosine hydroxylase: studies on the phosphorylation of a purified preparation of the brain enzyme by cyclic AMP-dependent protein kinase.J. Pharmacol. Exp. Ther. 216, 647–653.
Ehret M., Cash C. D., Hamon M., and Maitre M. (1989) Partial demonstration of the phosphorylation of rat brain tryptophan hydroxylase by Ca2+/ calmodulin-dependent protein kinase.J. Neurochem. 52, 1886–1891.
Friedman P. A., Kappelman A. H., and Kaufman S. (1972) Partial purification and characterization of tryptophan hydroxylase from rabbit hindbrain.J. Biol. Chem. 247, 4165–4173.
Fujisawa H. and Nakata H. (1987) Tryptophan 5-monooxygenase from mouse mastocytoma clone P815.Methods in Enzymol. 142, 93–96.
Furukawa Y., Ikuta N., Omata S., Yamauchi T., Isobe T., and Ichimura T. (1993) Demonstration of the phosphorylation-dependent interaction of tryptophan hydroxylase with the 14-3-3 protein.Biochem. Biophys. Res. Commun. 194, 144–149.
Grenett H. E., Ledley F. D., Reed L. L., and Woo S. L. C. (1987) Full-length cDNA for rabbit tryptophan hydroxylase: functional domains and evolution of aromatic amino acid hydroxylases.Proc. Natl. Acad. Sci. USA 84, 5530–5534.
Grima B., Lamouroux A., Blanot F., Biguet N. F., and Mallet J. (1985) Complete coding sequence of rat tyrosine hydroxylase mRNA.Proc. Natl. Acad. Sci. USA 82, 617–621.
Huang D. and Amero S. A. (1997) Measurement of antigen by enhanced chemiluminescent western blot.BioTechniques 22, 454–458.
Hufton S. E., Jennings I. G., and Cotton R. G. H. (1995) Structure and function of the aromatic amino acids hydroxylases.J. Biochem. 311, 353–366.
Jequier E., Robinson D. S., Lovenberg W., and Sjoerdsma A. (1969) Further studies on tryptophan hydroxylase in rat brainstem and beef pineal.Biochem. Pharmacol. 18, 1071–1081.
Johansen P. A., Jennings I. G., Cotton R. G. H., and Kuhn D. M. (1995) Tryptophan hydroxylase is phosphorylated by protein kinase A.J. Neurochem. 65, 882–888.
Johansen P. A., Jennings I. G., Cotton R. G. H., and Kuhn D. M. (1996) Phosphorylation and activation of tryptophan hydroxylase by exogenous protein kinase A.J. Neurochem. 66, 817–823.
Joh T. H., Park D. H., and Reis D. J. (1978) Direct phosphorylation of brain tyrosine hydroxylase by cyclic AMP-dependent protein kinase: mechanism of enzyme activation.Proc. Natl. Acad. Sci. USA 75, 4744–4746.
Joh T. H., Shikimi T., Pickel V. M., and Reis D. J. (1975) Brain tryptophan hydroxylase: purification of, production of antibodies to, and cellular and ultrastructural localization in serotonergic neurons of rat midbrain.Proc. Natl. Acad. Sci. USA 72, 3575–3579.
Kuhn D. M., Ruskin B., and Lovenberg W. (1980) Tryptophan hydroxylase: role of oxygen, iron, sulfhydryl groups as determinants of stability and catalytic activity.J. Biol. Chem. 255, 4137–4143.
Kuhn D. M., Arthur R. A. Jr., and States J. C. (1997) Phosphorylation and activation of brain tryptophan hydroxylase: identification of serine-58 as a substrate site for protein kinase A.J. Neurochem. 68, 2220–2223.
Kumer S. C., Mockus S. M., Rucker P. J., and Vrana K. E. (1997) Amino terminal deletion analysis of tryptophan hydroxylase: PKA phosphorylation occurs at serine-58.J. Neurochem., in press.
Kumer S. C. and Vrana K. E. (1996) The intricate regulation of tyrosine hydroxylase activity and gene expression.J. Neurochem. 67, 443–462.
Laemmli U. K. (1970) Cleavage of structural protein during the assembly of the head of bacteriophage T4.Nature 227, 680–685.
Ledley F. D., DiLella A. G., Kwok S. C. M., and Woo S. L. C. (1985) Homology between phenylalanine and tyrosine hydroxylase reveals common structural and functional domains.Biochemistry 24, 3389–3394.
Levitt M., Spector S., Sjoerdsma A., and Udenfriend S. (1965) Elucidation of the rate-limiting step in norepinephrine biosynthesis of the perfused guineapig heart.J. Pharmacol. Ther. 148, 1–7.
Liu X. and Vrana K. E. (1991) Leucine zippers and coiled-coils in the aromatic amino acid hydroxylases.Neurochem. Int. 18, 27–31.
Lohse D. L. and Fitzpatrick P. F. (1993) Identification of the intersubunit binding region in rat tyrosine hydroxylase.Biochem. Biophys. Res. Commun. 197, 1543–1548.
Makita Y., Okuno S., and Fujisawa H. (1990) Involvement of activator protein in the activation of tryptophan hydroxylase by cAMP-dependent protein kinase.FEBS Lett. 268, 185–188.
Mockus S. M., Kumer S. C., and Vrana K. E. (1997) Carboxyl terminal deletion analysis of tryptophan hydroxylase.Biochim. Biophys. Acta, in press.
Nakata H. and Fujisawa H. (1982a) Purification and properties of tryptophan 5-monoxygenase from rat brainstem.Eur. J. Biochem. 122, 41–47
Nakata H. and Fujisawa H. (1982b) Tryptophan 5-monoxygenase from mouse mastocytoma.Eur. J. Biochem. 124, 595–601.
Nukiwa T., Tohyama C., Okita T., and Ichiyama A. (1982) Purification and some properties of bovine pineal tryptophan 5-monooxygenase.Biochem. Biophys. Res. Commun. 60, 1029–1035.
O'Neill R. R., Mitchell L. G., Merril C. R., and Rasband W. S. (1989) Use of image analysis to quantitate changes in form of mitochondrial DNA after x-irradiation.Appl. Theor. Electrophor. 1, 163–167.
Ota A., Yoshida S., and Nagatsu T. (1995) Deletion mutagenesis of human tyrosine hydroxylase type 1 regulatory domain.Biochem. Biophys. Res. Commun. 213, 1099–1106.
Quinsey N. S., Lenaghan C. M., and Dickson P. W. (1996) Identification of Gln313 and Pro327 as residues critical for substrate inhibition in tyrosine hydroxylase.J. Neurochem. 66, 908–914.
Reinhard J., Smith G., and Nichol C. (1986) A rapid and sensitive assay for tyrosine-3-monooxygenase based upon the release of3H2O and adsorption of [3H] tyrosine by charcoal.Life Sci. 39, 2185–2189.
Ribeiro P., Wang Y., Citron B. A., and Kaufman S. (1993) Deletion mutagenesis of rat PC12 tyrosine hydroxylase regulatory and catalytic domains.J. Mol. Neurosci. 4, 125–139.
Sambrook J., Fritsch E. F., and Maniatis T. (1989)Molecular Cloning: A Laboratory Manual, 2nd ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
Tipper P. T., Citron B. A., Ribeiro P., and Kaufman S. (1994) Cloning and expression of rabbit and human brain tryptphan hydroxylase cDNA inEscherichia coli.Arch. Biochem. Biophys.,315, 445–453.
Vitto A. and Mandell A. J. (1981) Stability properties of activated tryptophan hydroxylase from rat midbrain.J. Neurochem. 37, 601–607.
Vrana S. L., Dworkin S. I., and Vrana K. E. (1993) Radioenzymatic assay for tryptophan hydroxylase: [3H]H2O release assessed by charcoal adsorption.J. Neurosci. Meth.,48, 123–129.
Vrana K. E., Rucker P. J., and Kumer S. C. (1994a) Recombinant rabbit tryptophan hydroxylase is a substrate for cAMP-dependent protein kinase.Life Sci. 55, 1045–1052.
Vrana K. E., Walker S. J., Rucker P., and Liu X. (1994b) A carboxyl terminal leucine zipper is required for tyrosine hydroxylase tetramer formation.J. Neurochem. 63, 2014–2020.
Vulliet P. R., Langan T. A., and Weiner N. (1980) Tyrosine hydroxylase: a substrate of cyclic AMP-dependent protein kinase.Proc. Natl. Acad. Sci. USA 77, 92–96.
Walker S. J., Liu X., Roskoski R. Jr., and Vrana K. E. (1994) Catalytic core of rat tyrosine hydroxylase: terminal deletion analysis of a bacterially-expressed enzyme.Biochim. Biophys. Acta 1206, 113–119.
Wang Y. H., Citron B., Ribeiro P., and Kaufman S. (1991) High-level expression of rat PC12 tyrosine hydroxylase cDNA inE. coli: purification and characterization of the cloned enzyme.Proc. Natl. Acad. Sci. USA 88, 8779–8783.
Widmer F., Mutus, B., RamaMurthy J., Snieckus V. A., and Viswanatha, T. (1975) Partial purification of rabbit hind brain tryptophan hydroxylase by affinity chromatography.Life Sci. 17, 1297–1302.
Yamauchi T. and Fujisawa H. (1979) Regulation of bovine adrenal tyrosine-3-monooxygenase by phosphorylation-dephosphorylation reaction catalyzed by adenosine 3′,5′-monophosphate-dependent protein kinase and phosphoprotein phosphatase.J. Biol. Chem. 254, 6408–6413.
Yang X. J. and Kaufman S. (1994) High-level expression and deletion mutagenesis of human tryptophan hydroxylase.Proc. Natl. Acad. Sci. USA 91, 6659–6663.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Mockus, S.M., Kumer, S.C. & Vrana, K.E. A chimeric tyrosine/tryptophan hydroxylase. J Mol Neurosci 9, 35–48 (1997). https://doi.org/10.1007/BF02789393
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF02789393