Abstract
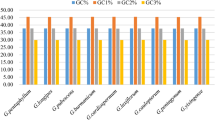
A detailed comparison was made of codon usage of chloroplast genes with their host (nuclear) genes in the four angiosperm speciesOryza sativa, Zea mays, Triticum aestivum andArabidopsis thaliana. The average GC content of the entire genes, and at the three codon positions individually, was higher in nuclear than in chloroplast genes, suggesting different genomic organization and mutation pressures in nuclear and chloroplast genes. The results of Nc-plots and neutrality plots suggested that nucleotide compositional constraint had a large contribution to codon usage bias of nuclear genes inO. sativa, Z. mays, andT. aestivum, whereas natural selection was likely to be playing a large role in codon usage bias in chloroplast genomes. Correspondence analysis and chi-test showed that regardless of the genomic environment (species) of the host, the codon usage pattern of chloroplast genes differed from nuclear genes of their host species by their AU-richness. All the chloroplast genomes have predominantly A- and/or U-ending codons, whereas nuclear genomes have G-, C- or U-ending codons as their optimal codons. These findings suggest that the chloroplast genome might display particular characteristics of codon usage that are different from its host nuclear genome. However, one feature common to both chloroplast and nuclear genomes in this study was that pyrimidines were found more frequently than purines at the synonymous codon position of optimal codons.
Similar content being viewed by others
References
Bellgard M., Schibeci D., Trifonov E. and Gojobori T. 2001 Early detection of G + C differences in bacterial species inferred from the comparative analysis of the two completely sequencedHelicobacter pylori strains.J. Mol. Evol. 53, 465–468.
Bernardi G. 1993 The isochore organization of the human genome and its evolutionary history—a review.Gene 135, 57–66.
Bulmer M. 1988 Are codon usage patterns in unicellular organisms determined by selectionmutation balance?J. Mol. Biol. 1, 15–26.
Campbell W. H. and Gowri G. 1990 Codon usage in higher plants, green algae, and cyanobacteria.Plant Physiol. 92, 1–11.
Chiapello H., Lisacek F., Caboche M. and Hénaut A. 1998 Codon usage and gene function are related in sequences ofArabidopsis thaliana.Gene 209, GC1-GC38.
de Amicis F. and Marchetti S. 2000 Intercodon dinucleotides affect codon choice in plant genes.Nucleic Acids Res. 28, 3339–3345.
Duret L. 2000 tRNA gene number and codon usage in theC. elegans genome are coadapted for optimal translation of highly expressed genes.Trends Genet. 16, 287–289.
Duret L. and Mouchiroud D. 1999 Expression pattern and, surprisingly, gene length shape codon usage inCaenorhabditis, Drosophila, andArabidopsis.Proc. Natl. Acad. Sci. USA 96, 4482–4487.
Fennoy S. L. and Bailey-Serres J. 1993 Synonymous codon usage inZea mays L. nuclear genes is varied by levels of C and Gending codons.Nucleic Acids Res. 21, 5294–5300.
Fernandez V., Zavala A. and Musto H. 2001 Evidence for translational selection in codon usage inEchinococcus spp.Parasitology 123, 203–209.
Grantham R., Gautier C. and Gouy M. 1980 Codon frequencies in 119 individual genes confirm consistent choices of degenerate bases according to genome type.Nucleic Acids Res. 8, 1893–1912.
Greenacre M. J. 1984Theory and applications of correspondence analysis. Academic Press, London.
Grocock R. J. and Sharp P. M. 2002 Synonymous codon usage inPseudomonas aeruginosa PA01.Gene 289, 131–139.
Gu W., Zhou T., Ma J., Sun X. and Lu Z. 2004. The relationship between synonymous codon usage and protein structure inEscherichia coli andHomo sapiens.BioSystems 73, 89–97.
Gupta S. K., Bhattacharyya T. K. and Ghosh T. C. 2004 Synonymous codon usage inLactococcus lactis: mutational bias versus translational selection.J. Biomol. Struct. Dyn. 21, 1–9.
Hou Z. C. and Yang N. 2003 Factors affecting codon usage inYersinia pestis.Acta Biochimica et Biophysica Sinica 35, 580–586.
Iannacone R., Grieco P. D. and Cellini F. 1997 Specific sequence modifications of a cry3B endotoxin gene result in high levels of expression and insect resistance.Plant Mol. Biol. 34, 485–496.
Ikemura T. 1985 Codon usage and tRNA content in unicellular and multicellular organisms.Mol. Biol. Evol. 2, 13–34.
Karlin S. and Mrázek J. 1996 What drives codon choices in human genes?J. Mol. Biol. 262, 459–472.
Kawabe A. and Miyashita N. T. 2003 Patterns of codon usage bias in three dicot and four monocot plant species.Genes Genet. Syst. 78, 343–352.
Kikuchil S., Satoh K., Nagata T., Kawagashira N., Doi K., Kishimoto N., Yazaki J., Ishikawa M., Yamada H. and Ooka H. 2003 Collection, mapping, and annotation of over 28000 cDNA clones fromjaponica rice.Science 301, 376–379.
Lerat E., Capy P. and Biémont C. 2002 Codon usage by transposable elements and their host genes in five species.J. Mol. Evol. 54, 625–637.
Liu Q. P., Feng Y., Zhao X., Dong H. and Xue Q. Z. 2004 Synonymous codon usage bias inOryza sativa.Plant Sci. 167, 101–105.
Moriyama E. N. and Powell J. R. 1998 Gene length and codon usage bias inDrosophila melanogaster, Saccharomyces cerevisiae andEscherichia coli.Nucleic Acids Res. 26, 3188–3193.
Morton B. R. 1998 Selection on the codon bias of chloroplast and cyanelle genes in different plant and algal lineages.J. Mol. Evol. 46, 449–459.
Morton B. R. 1999 Strand asymmetry and codon usage bias in the chloroplast genome ofEuglena gracilis.Proc. Natl. Acad. Sci. USA 96, 5123–5128.
Morton B. R. 2003 The role of contextdependent mutations in generating compositional and codon usage bias in grass chloroplast DNA.J. Mol. Evol. 56, 616–629.
Murray E. E., Lotzer J. and Eberle M. 1989 Codon usage in plant genes.Nucleic Acids Res. 17, 477–498.
Musto H., Cruveiller S., Onofrio G. D., Romero H. and Bernardi G. 2001 Translational selection on codon usage inXenopus laevis.Mol. Biol. Evol. 18, 1703–1707.
Naya H., Romero H., Carels N., Zavala A. and Musto H. 2001 Translational selection shapes codon usage in the GCrich genomes ofChlamydomonas reinhardtii.FEBS Lett. 501, 127–130.
Peixoto L., Zavala A., Romero H. and Musto H. 2003 The strength of translational selection for codon usage varies in the three replicons ofSinorhizobium melioti.Gene 320, 109–116.
Percudani R., Pavesi A. and Ottonello S. 1997 Transfer RNA gene redundancy and translational selection inSaccharomyces cerevisiae.J. Mol. Biol. 268, 322–330.
Romero H., Zavala A. and Musto H. 2000 Codon usage inChlamydia trachomatis is the result of strandspecific mutational biases and a complex pattern of selective forces.Nucleic Acids Res. 28, 2084–2090.
Romero H., Zavala A., Musto H. and Bernardi G. 2003 The influence of translational selection on codon usage in fishes from the family Cyprinidae.Gene 317, 141–147.
Rouwendal G. J. A., Mendes O., Wolbert E. J. H. and de Boer A. D. 1997 Enhanced expression in tobacco of the gene encoding green fluorescent protein by modification of its codon usage.Plant Mol. Biol. 33, 989–999.
Salinas J., Matassi G., Montero L. M. and Bernardi G. 1988 Compositional compartmentalization and compositional patterns in the nuclear genomes of plants.Nucleic Acids Res. 16, 4269–4285.
Sharp P. M. and Li W. H. 1986 An evolutionary perspective on synonymous codon usage in unicellular organisms.J. Mol. Evol. 24, 28–38.
Sharp P. M. and Matassi G. 1994 Codon usage and genome evolution.Curr. Opin. Genet. Dev. 4, 851–860.
Sharp P. M., Stenico M., Peden J. F. and Lloyd A. T. 1993 Codon usage: mutational bias, translational selection, or both?Biochem. Soc. Trans. 21, 835–841.
Sharp P. M., Tuohy T. M. and Mosurski K. R. 1986 Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes.Nucleic Acids Res. 14, 5125–5143.
Shi X. F., Huang J. F., Liang C. R., Liu S. Q., Xie J. and Liu C. Q. 2001 Is there a close relationship between synonymous codon bias and codonanticodon binding strength in human genes?Chinese Sci. Bulletin 12, 1015–1019.
Shields D. C., Sharp P. M., Higgins D. G. and Wright F. 1988 “Silent” sites inDrosophila genes are not neutral: evidence of selection among synonymous codons.Mol. Biol. Evol. 5, 704–716.
Singer G. A. C. and Hickey D. A. 2003 Thermophilic prokaryotes have characteristic patterns of codon usage, amino acid composition and nucleotide content.Gene 317, 39–47.
Stenico M., Lloyd A. T. and Sharp P. M. 1994 Codon usage inCaenorhabditis elegans: delineation of translational selection and mutational biases.Nucleic Acids Res. 22, 2437–2446.
Sueoka N. 1988 Directional mutation pressure and neutral molecular evolution.Proc. Natl. Acad. Sci. USA 85, 2653–2657.
Sugiura M. 1992 The chloroplast genome.Plant Mol. Biol. 19, 149–168.
Wright F. 1990 The “effective number of codons” used in a gene.Gene 87, 23–29.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Liu, Q., Xue, Q. Comparative studies on codon usage pattern of chloroplasts and their host nuclear genes in four plant species. J Genet 84, 55–62 (2005). https://doi.org/10.1007/BF02715890
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1007/BF02715890