Abstract
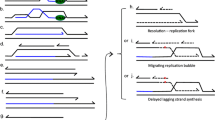
Stationary-phase mutagenesis in nondividingE. coli cells exposed to a nonlethal stress was, a few years ago, claimed to be a likely case of a Lamarckian mechanism capable of producing exclusively useful mutations in a directed manner. After a heated debate over the last decade it now appears to involve a Darwinian mechanism that generates a transient state of hypermutagenesis, operating on a large number of sites spread over the entire genome, at least in a proportion of the resting cells. Most of the studies that clarified this position were on the reversion of a frameshift mutation present in alacI-lacZ fusion inE. coli strain FC40. Several groups have extensively examined both the sequence changes associated with these reversions and the underlying genetic requirements. On the basis of our studies on the genomic sequence analysis, we recently proposed a model to explain the specific changes associated with the reversion hotspots. Here we propose a more detailed version of this model that also takes into account the observed genetic requirements of stationary-state mutagenesis. Briefly, G:T/U mismatches produced at methylatable cytosines are preferentially repaired in nondividing cells by the very short patch mismatch repair (VSPMR) mechanism which is itself mutagenic and can produce mutations in very short stretches located in the immediate vicinity of these cytosine methylation sites. This mechanism requires a homologous or homeologous strand invasion step and an error-prone DNA synthesis step and is dependent on RecA, RecBCD and a DNA polymerase. The process is initiated near sequences recognized by Dcm and Vsr enzymes and further stimulated if these sequences are a part of CHI or CHI-like sequences, but a double-strand-break-dependent recombination mediated by the RecBCD pathways proposed by others seems to be nonessential. The strand transfer step is proposed to depend on RecA, RuvA, RuvB and RuvC and is opposed by RecG and MutS. The model also gives interesting insights into the evolution of theE. coli genome.
Similar content being viewed by others
References
Bhattacharjee S. K. and Mahajan S. K. 1998 The origin of adaptive mutations: explaining the mutational spectra of Lac+ revertants of theEscherichia coli strain FC40.Curr. Sci. 74, 583–590.
Blattner F. R., Plunkett G. III, Bloch C. A., Perna N. T., Burland V. and Riley M. 1997 The complete genome sequence ofEscherichia coli K-12.Science 277, 1453–1474.
Boe L. 1990 Mechanism for induction of adaptive mutations inEscherichia coli.Mol. Microbiol. 4, 597–601.
Brégeon D., Matic I., Radman M. and Taddei F. 1999 Inefficient mismatch repair: genetic defects and down regulation.J. Genet. 78, 21–28.
Cairns J. and Foster P. L. 1991 Adaptive reversions of a frameshift mutation inEscherichia coli.Genetics 128, 695–701.
Cairns J., Overbaugh J. and Miller S. 1988 The origin of mutants.Nature 335, 142–145.
Cascalho M., Wong J., Steinberg C. and Wabl M. 1998 Mismatch repair co-opted by hypermutation.Science 279, 1207–1210.
Colbert T., Taylor A. F. and Smith G. R. 1998 Genomics, Chi sites and codons: ‘islands of preferred DNA pairing’ are oceans of ORFs.Trends Genet. 14, 485–488.
Escarceller M., Hicks J., Gudmundsson G., Trump G., Touati D., Lovett S., Foster P. L., McEntee K. and Goodman M. F. 1994 Involvement ofEscherichia coli DNA polymerase II in response to oxidative damage and adaptive mutation.J. Bacteriol. 176, 6221–6228.
Feng G., Tsui H. T. and Winkler M. E. 1996 Depletion of the cellular amounts of the MutS and MutH methyl-directed mismatch repair proteins in stationary-phaseEscherichia coli K-12 cells.J. Bacteriol. 178, 2388–2396.
Fijalkowska I. J., Dunn R. I. and Schaaper R. M. 1993 Mutation ofEscherichia coli with increased fidelity of DNA replication.Genetics 134, 1023–1030.
Foster P. L. 1993 Adaptive mutation: the uses of adversity.Annu. Rev. Microbiol. 47, 467–504.
Foster P. L. 1997 Nonadaptive mutations occur on the F’ episome during adaptive mutation conditions inEscherichia coli.J. Bacteriol. 179, 1550 -1554.
Foster P. L. 1999a Mechanisms of stationary phase mutation: a decade of adaptive mutation.Annu. Rev. Genet. 33, 57–88.
Foster P. L. 1999b Are adaptive mutations due to a decline in mismatch repair? The evidence is lacking.Mutat. Res. 436, 179–184.
Foster P. L. and Cairns J. 1992 The mechanisms of directed mutation.Genetics 131, 783–789.
Foster P. L. and Cairns J. 1998 Adaptive mutation of alacZ amber allele.Genetics 150, 1329–1330.
Foster P. L. and Rosche W. A. 1998 Levels of the Vsr endonuclease do not regulate stationary-phase reversion of a Lac- frameshift allele inEscherichia coli.J. Bacteriol. 180, 1944 -1946.
Foster P. L. and Rosche W. A. 1999a Adaptive mutations inEscherichia coli strain FC40.J. Genet. 78, 7–11.
Foster P. L. and Rosche W. A. 1999b Increased episomal replication accounts for the high rate of adaptive mutations inrecD mutants ofEscherichia coli.Genetics 152, 15–30.
Foster P. L. and Trimarchi J. M. 1994 Adaptive reversion of a frameshift mutation inEscherichia coli by simple base deletions in homopolymeric runs.Science 265, 407–409.
Foster P. L. and Trimarchi J. M. 1995 Adaptive reversion of an episomal frameshift mutation inEscherichia coli requires conjugal functions but not actual conjugation.Proc. Natl. Acad. Sci. USA 92, 5487–5490.
Foster P. L., Gudmundsson J. M., Trimarchi J. M., Cai H. and Goodman M. F. 1995 Proofreading-defective DNA polymerase II increases adaptive mutation inEscherichia coli.Proc. Natl. Acad. Sci. USA 92, 7951–7955.
Foster P. L., Trimarchi J. M. and Maurer R. A. 1996 Two enzymes, both of which process recombination intermediates, have opposite effects on adaptive mutation inEscherichia coli.Genetics 142, 25–37.
Galitski T. and Roth J. R. 1995 Evidence that F’ transfer replication underlies apparent adaptive mutation.Science 268, 421–423.
Hall B.G. 1999 Spectra of spontaneous growth-dependent and adaptive mutations atebgR.J. Bacteriol. 181, 1149–1155.
Harris R. S., Longerich J. and Rosenberg S. M. 1994 Recombination in adaptive mutation.Science 264, 258–260.
Harris R. S., Ross K. J. and Rosenberg S. M. 1996 Opposing roles of the Holliday junction processing systems ofEscherichia coli in recombination dependent adaptive mutation.Genetics 142, 681–691.
Harris R. S., Bull H. J. and Rosenberg S. M. 1997a A direct role for DNA polymerase III in adaptive reversion of a frameshift mutation inEscherichia coli.Mutat. Res. 375, 19–24.
Harris R. S., Feng G., Ross K. J., Sidhu R., Thulin C., Longerich S., Szigety S. K., Hastings P. J., Winkler M. E. and Rosenberg S. M. 1997b Mismatch repair protein MutL becomes limiting during stationary-phase mutation.Genes Dev. 11, 2426–2437.
Harris R. S., Feng G., Ross K. J., Sidhu R., Thulin C., Longerich S., Szigety S. K., Hastings P. J., Winkler M. E. and Rosenberg S. M. 1999 Mismatch repair is diminished during stationary phase mutations.Mutat. Res. 437, 51–60.
Hennecke F., Kolmar H., Brandi K. and Fritz H.-J. 1991 Thevsr gene product ofE. coli K-12 is a strand and sequence specific DNA mismatch endonuclease.Nature 353, 776–778.
Jayaraman R. 1992 Cairnsian mutagenesis inEscherichia coli: Genetic evidence for two pathways regulated bymutS andmutL genes.J. Genet. 71, 23–41.
Karin D., Alexander A., Joachim F.-H. and Marinus M. G. 1998 TheEscherichia coli MutL protein stimulates binding of Vsr and MutS to heterologous DNA.Nucl. Acids Res. 26, 948–953.
Kuzminov A. 1995 Collapse and repair of replication fork inEscherichia coli.Mol. Microbiol. 16, 373–384.
Lombardo M.-J. and Rosenberg S. M. 1999 Hypermutation in stationary-phaseE. coli: tales from thelac operon.J. Genet. 78, 13–20.
Longerich S., Galloway A. M., Harris R. S., Wong C. and Rosenberg S. M. 1995 Adaptive mutation sequences reproduced by mismatch repair deficiency.Proc. Natl. Acad. Sci. USA 92, 12017–12020.
McEnzie G. J., Lombardo M. J. and Rosenberg S. M. 1998 Recombination-dependent mutations inE. coli occur in stationary phase.Genetics 149, 1163–1165.
McMilin K. D., Stahl M. M. and Stahl F. W. 1974 Rec-mediated recombinational hotspot activity in bacteriophage lambda. I. Hotspot activity associated with Spideletions and bio substitutions.Genetics 77, 409–423.
Mahajan S. K. 1988 Pathways of homologous recombination inEscherichia coli K-12. InGenetic recombination (ed. R. Kucherlapati and G. R. Smith), pp. 87–128. American Society for Microbiology, Washington, DC.
Mahajan S. K., Shirke N. D. and Bhattacharjee S. K. 1998 The possible involvement of CHI sequences in adaptive mutagenesis: evidence from sequence analysis.J. Genet. 77, 105–114.
Martin A. 1997 Studies with enzymes of recombination pathways inEscherichia coli K-12. Ph.D. thesis, University of Mumbai, Mumbai, India.
Massey R. C., Rainey P. B., Sheehan B. J., Keane O. M. and Dorman C. J. 1999 Environmentally constrained mutation and adaptive evolution inSalmonella.Curr. Biol. 9, 1477–1480.
Modrich P. 1991 Mechanisms and biological effects of mismatch repair.Annu.Rev. Genet. 25, 229–253.
Modrich P. and Lahue R. 1996 Mismatch repair in replication fidelity, genetic recombination and cancer biology.Annu. Rev. Biochem. 65, 101–133.
Peters J. E. and Benson S. A. 1995 Redundant transfer of F’ plasmids occurs betweenEscherichia coli cells during nonlethal selection.J. Bacteriol. 177, 847–850.
Privai M. J. and Cebula T. A. 1992 Sequence analysis of mutations arising during prolonged starvation ofSalmonella typhimurium.Genetics 132, 303–310.
Privai M. J. and Cebula T. A. 1996 Adaptive mutations and slowgrowing revenants ofEscherichia coli lacZ amber mutation.Genetics 144, 1337–1344.
Radicella J. P., Park P. U. and Fox M. S. 1995 Adaptive mutation inEscherichia coli: a role for conjugation.Science 268, 418–420.
Radman M. 1988 Mismatch repair and genetic recombination. InGenetic recombination (ed. R. Kucherlapati and G. R. Smith), pp. 169–192. American Society for Microbiology, Washington, DC.
Rangarajan S., Gudmundsson G., Qiu Z., Foster P. L. and Goodman M. F. 1997Escherichia coli DNA polymerase II catalyzes chromosomal and episomal DNA synthesisin vivo.Proc. Natl. Acad. Sci. USA 94, 946–951.
Reddy M. and Gowrishankar J. 1997 A genetic strategy to demonstrate the occurrence of spontaneous mutations in nondividing cells within colonies ofEscherichia coli.Genetics 147, 991–1001.
Rosenberg S. M., Longerich S., Gee P. and Harris R. S. 1994 Adaptive mutation by deletions in small mononucleotide repeats.Science 265, 405–407.
Shapiro J. A. 1984 Observation in the formation of clones containing araB-lacZ cistron fusion.Mol. Gen. Genet. 194, 79–90.
Torkelson J., Harris R., Lombardo M.-J., Nagendran C., Thulin C. and Rosenberg S. M. 1997 Genome wide hypermutations in a subpopulation of stationary-phase cells underlies recombination dependent adaptive mutation.EMBO J. 16, 3303–3311.
Tracy R. B., Chedin F. and Kowalczykowski S. C. 1997 The recombination hot spot Chi is embedded within islands of preferred DNA pairing sequences in theE. coli genome.Cell 90, 205–206.
Tsui H.-C, Feng G. and Winkler M. E. 1997 Negative regulation ofmutS andmutH repair gene expression by the Hfq and RpoS global regulators ofE. coli K-12.J. Bacteriol. 179, 7476–7487.
mWalker 1985 Inducible DNA repair systems.Annu. Rev. Biochem. 54, 425–457.
West S. C. 1996 The RuvABC and Holliday junction processing inEscherichia coli.J. Bacteriol. 178, 1237–1241.
Whitby M. C. and Lloyd R. G. 1995 Branch migration of three strand recombination intermediates by RecG, a possible pathway for securing exchanges initiated by 3′-tailed duplex DNA.EMBO J. 14, 3302–3310.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mahajan, S.K., Narayana Rao, A.V.S.S. & Bhattacharjee, S.K. Stationary-state mutagenesis inEscherichia coli: A model. J Genet 79, 1–7 (2000). https://doi.org/10.1007/BF02715869
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF02715869