Abstract
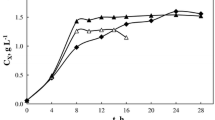
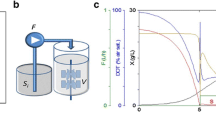
In our previous work (Xie and Wang, 1994a), a simplified stoichiometric model on energy metabolism for animal cell cultivation was developed. Fed-batch experiments were performed in T-flasks using this model in supplemental medium design (Xie and Wang, 1994b). In this work, the major pathways of glucose and glutamine metabolism were incorporated into the stoichiometric model. Fed-batch culture was conducted in a 2-liter bioreactor with appropriate process control strategies. Nutrient concentrations, especially glucose and glutamine, were maintained at constant but low levels through the automated feeding of a supplemental medium formulated using the improved stoichiometric model. The formation of toxic byproducts, such as ammonia and lactate (Hassellet al., 1991), was greatly reduced. The specific lactate production rate was decreased by 62-fold compared with batch culture in bioreactor and by 8-fold compared to fed-batch culture in T-flask using the previous stoichiometric model. Ammonia formation was also decreased compared with both the batch and fed-batch cultures. Most importantly, the monoclonal antibody concentration reached 900 mg l−1, an increase of 17- and 1.6-fold compared with the batch and fed-batch cultures respectively.
Similar content being viewed by others
Abbreviations
- Camm, n, Camm, n−1 :
-
ammonia concentration in the n and (n−1)th samples respectively
- mM; C samm :
-
ammonia concentration in the supplemental medium, mM
- CAb, n, CAb, n−1 :
-
antibody concentration in the n and (n−1)th samples respectively, mg l−1
- Ci :
-
concentration of the ith nutrient in the supplemental medium, mM
- Clac, n, Clac, n−1 :
-
lactate concentration in the n and (n−1)th samples respectively, mM
- C sk :
-
concentration of glucose or glutamine in the supplemental medium, mM
- Ck, n, Ck, n−1 :
-
concentration of glucose or glutamine in the n or(n−1)th sample respectively
- mM; Ct :
-
total concentration of glucose, amino acids, and vitamins in the supplemental medium, mM
- F1, F2, F3, F4, F5, F6, F7, F8, F9, F10, F11, F12 :
-
fluxes described in Fig. 1, mmole cell−1
- m:
-
total number of samples
- Nt :
-
total cell number in the reactor at culture time t, number of cells
- Nt, o :
-
total cell number in the reactor at the beginning of culture, number of cells
- Nt, n, Nt, n−1 :
-
total cells in the reactor when the n and (n−1)th samples were taken, respectively, number of cells
- ΔNt, n, ΔNt, m :
-
total cells produced since the initiation when the nth sample and the mth sample were taken, respectiveky, number of cells
- Nv :
-
viable cell number in the reactor at culture time t, number of cells
- Nv, n, Nv, n−1 :
-
viable cells in the reactor when the n and (n−1)th samples were taken, respectively, number of cells
- PAb, n :
-
monoclonal antibody produced between the n and (n−1) samples, mg
- Pamm, n :
-
ammonia produced between the n and (n−1) samples, mmole
- Pi, n :
-
amount of lactate or ammonia produced between the n and (n−1)th samples, mmole
- Plac, n :
-
lactate produced between the n and (n−1) samples, mmole
- P/O:
-
number of ATP molecules generated per NADH molecule oxidized
- qi :
-
specific production rate of ammonia, lactate, or antibody respectively, mmole cell−1 h−1 (for ammonia and lactate), and mg cell−1 h−1 (for antibody)
- t:
-
culture time, h
- tm :
-
culture time when the mth sample (last sample) was taken, h
- tn, tn−1 :
-
culture time when the n and (n−1)th samples were taken respectively, h
- ΔVF :
-
volume of supplemental medium fed to the reactor since the nth sample was taken, l
- ΔVV, n :
-
volume fed to the reactor between the (n−1)th and nth samples, l
- Vn, Vn−1 :
-
total volume in the reactor after the nth or (n−1)th sample were taken respectively, l
- Vs, j :
-
volume of the jth sample, l
- Vs, n, Vs, n−1 :
-
volume of the n and (n−1)th samples respectively, l
- Xt, j :
-
total cell density in the jth sample, cells l−1
- Xt, n :
-
total cell density of the nth sample, cells l−1
- Xv, n, Xv, n−1 :
-
viable cell density in the n and (n−1)th samples respectively, cells l−1
- Yamm/gln :
-
ratio of ammonia production to glucose consumption, mmol mmol−1
- Yi/cell :
-
ratio of the total ammonia or lactate production to the total production of cells, mmole cell−1
- Ylac/glc :
-
ratio of lactate production to glucose consumption, mmol mmol−1
- α:
-
specific death rate, h−1
- β:
-
total stoichiometric coefficient of glucose, amino acids, and vitamins, mmole cell−1
- δglc, n, δgln, n :
-
Amount of glucose or glutamine crespectively, consumed between the n and the (n−1)th sample, mmole
- δk, n :
-
Amount of glucose or glutamine consumed between the n and the (n−1)th sample, mmole
- μ:
-
specific growth rate, h−1
- θala, θasn, θasp, θglc, θglu, θgln, θgly, θpro, θser :
-
stoichiometric coefficient of alanine, asparagine, aspartate, glucose, glutamate, glutamine, glycine, proline, and serine respectively including energy metabolism and biosynthesis of nonessential amino acids, mmole cell−1
- θ cmala , θ cmasn , θ cmasp , θ cmglc , θ cmglu , θ cmgln , θ cmgly , θ cmpro , θ cmser :
-
stoichiometric coefficient of alanine, asparagine, aspartate, glucose, glutamate, glutamine, glycine, proline, and serine respectively in the biosyntheses of cell mass and product, excluding energy metabolism and synthesis from glutamine, mmole cell−1
- θATP :
-
stoichiometric coefficient for ATP in syntheses of cell mass and product, mmole cell−1
- θ englc :
-
stoichiometric coefficient of glucose in energy metabolism, mmol cell−1
- θi :
-
stoichiometric coefficient of nutrient, mmol cell−1
- θ cmi :
-
stoichiometric coefficient of nutrient in cell mass and product formation without consideration of consumption in energy formation and the production of nonessential amino acids from glutamine, mmol cell−1
- τ:
-
integration of viable cells over culture time, cells h
- cm:
-
cell mass (include product) syntheses
- en:
-
energy metabolism
- s:
-
supplemental medium
- Ab:
-
antibody
- ala:
-
alanine
- amm:
-
ammonia
- asn:
-
asparagine
- asp:
-
aspartate
- F:
-
feeding
- glc:
-
glucose
- gln:
-
glutamine
- glu:
-
glutamate
- gly:
-
glycine
- i:
-
index for nutrients, index for ammonia or lactate
- j:
-
index of sample
- k:
-
stands for glucose or glutamine
- lac:
-
lactate
- m:
-
total number of sample
- n:
-
index of sample
- p:
-
product
- pro:
-
proline
- ser:
-
serine
- t:
-
time or total cells
- v:
-
vitamin or viable cells
References
Adamson SR, Fitzpatrick SL and Behie LA (1983) In vitro production of high titer monoclonal antibody by hybridoma cells in dialysis culture. Biotechnol. Lett. 5: 573–578.
Bligh EG and Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917.
Fike R, Kubiak J, Price P and Jayme D (1993) Feeding strategies for enhanced hybridoma productivity: automated concentrate supplementation. BioPharm. 6: 49–54.
Folch J, Lees M and Sloane Stanley GH (1957) A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226: 497–509.
Gerschenson LE, Mead JF, Harary I and Haggerty Jr. DF (1967) Studies on the essential fatty acids on growth rate, fatty acid composition, oxidative phosphorylation and respiratory control of HeLa cells in culture. Biochim. Biophys. Acta 131: 42–49.
Glasken MW, Huang C and Sinskey AJ (1989) Mathematical descriptions of hybridoma culture kinetics. III. simulation of fedbatch bioreactors. J. Biotechnol. 10: 39–66.
Glacken MW (1987) Development of mathematical descriptions of mammalian cell culture kinetics for the optimization of fed-batch bioreactors. Ph. D. thesis, MIT, Cambridge, MA, USA
Hanson RS and Philips JA (1981) Chemical Composition. In: Gerhardt P et al. (eds.). Manual of Methods for General Bacteriology (pp. 329–364) American Society for Microbiology, Washington, D.C.
Hassell T, Gleave S and Butler M (1991) Growth inhibition in animal cell culture: The effect of lactate and ammonia. Applied Biochem. Biotechnol. 30: 29–41.
Hu WS, Dodge TC, Frame KK and Himes VB (1987) Effect of glucose on the cultivation of mammalian cells. Develop. Biol. Stand. 66: 279–290.
Linardos TI, Kalogerakis N and Behie LA (1991) The effect of specific growth rate and death rate on monoclonal antibody production in hybridoma chemostat cultures. Can. J. Chem. Eng. 69: 429–438.
Linardos TI, Kalogerakis N and Behie LA (1992) Monoclonal antibody production in dialyzed continuous suspension culture. Biotechnol. Bioeng. 39: 504–510.
Lindell PI (1992) Dynamic operation of mammalian cell fed-batch bioreactors. Ph.D. thesis, MIT, Cambridge, MA, USA.
Luan YT, Mutharasan R and Magee WE (1987) Strategies to extend the longevity of hybridomas in culture and promote yield of monoclonal antibodies. Biotechnol. Lett. 9: 691–696.
Nelson GJ (1975) Isolation and purification of lipids from animal tissues. In: Perkins EG (ed.) Analysis of lipids and lipoproteins (pp. 1–22) American Oil Chemists Society, Champaign, Illinois.
Oh SKW, Vig P, Chua F, Teo WK Yap MGS (1993) Substantial overproduction of antibodies by applying osmotic pressure and sodium butyrate. Biotechnol. Bioeng. 42: 601–610.
Ozturk S.S and Palsson BO (1991) Effect of medium osmolarity on hybridoma growth, metabolism, and antibody production. Biotechnol. Bioeng. 37: 989–993.
Packer L (1967) Experiments in cell physiology. Academic press, New York.
Pendse GJ and Bailey JE (1990) Effects of growth factors on cell proliferation and monoclonal antibody production of batch hybridoma cultures. Biotechnol. Lett. 12: 487–492.
Read SM (1984) Techniques in proteins and enzyme biochemistry, part I supplement: techniques for determining protein concentration. In: Tipton KF (ed.) Techniques in the life sciences. (pp. 1–34) Elsevier Scientific, New York.
Stein J and Smith G (1982) Techniques in lipid and membrane biochemistry, part I: extraction methods. In: Hesketh TR et al. (eds.) Techniques in the life sciences. (pp. 1–10) Elsevier/North-Holland Scientific, New York.
Xie L and Wang DIC (1994a) Stoichiometric analysis of animal cell growth and its application of medium design. Biotechnol. Bioeng. 43: 1164–1174.
Xie L and Wang DIC (1994b) Fed-batch cultivation of animal cells using different medium design concepts and feeding strategies. Biotechnol. Bioeng. 43: 1175–1189.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Xie, L., Wang, D.I.C. Applications of improved stoichiometric model in medium design and fed-batch cultivation of animal cells in bioreactor. Cytotechnology 15, 17–29 (1994). https://doi.org/10.1007/BF00762376
Issue Date:
DOI: https://doi.org/10.1007/BF00762376