Abstract
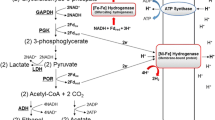
In Alcaligenes eutrophus H16 a pleiotropic DNA-region is involved in formation of catalytically active hydrogenases. This region lies within the hydrogenase gene cluster of megaplasmid pHG1. Nucleotide sequence determination revealed five open reading frames with significant amino acid homology to the products of the hyp operon of Escherichia coli and other hydrogenase-related gene products of diverse organisms. Mutants of A. eutrophus H16 carrying Tn5 insertions in two genes (hypB and hypD) lacked catalytic activity of both soluble (SH) and membrane-bound (MBH) hydrogenase. Immunological analysis showed that the mutants contained SH-and MBH-specific antigen. Growing the cells in the presence of 63Ni2+ yielded significantly lower nickel accumulation rates of the mutant strains compared to the wild-type. Analysis of partially purified SH showed only traces of nickel in the mutant protein suggesting that the gene products of the pleiotropic region are involved in the supply and/or incorporation of nickel into the two hydrogenases of A. eutrophus.
Similar content being viewed by others
References
Bourne HR, Sanders DA, McCormick R (1991) The GTPase superfamily: conserved structure and molecular mechanism. Nature 349: 117–127
Bruschi M, Guerlesquin F (1988) Structure, function and evolution of bacterial ferredoxins. FEMS Microbiol Rev 54: 155–176
Chen JC, Mortenson LE (1992) Identification of six open reading frames from a region of the Azotobacter vinelandii genome likely involved in dihydrogen metabolism. Biochim Biophys Acta 1131: 199–202
Colbeau A, Richaud P, Toussaint B, Caballero J, Elster C, Delphin C, Smith RL, Chabert J, Vignais PM (1993) Organization of the genes necessary for hydrogenase expression in Rhodobacter capsulatus. Sequence analysis. Identification of two hyp regulatory mutants. Mol Microbiol (in press)
Cole ST (1987) Nucleotide sequence and comparative analysis of the frd operon encoding the fumarate reductase of Proteus vulgaris. Eur J Biochem 167: 481–488
Cussac V, Ferrero RL, Labigne A (1992) Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogenlimiting conditions. J Bacteriol 174: 2466–2473
Du L, Stejskal F, Tibelius KH (1992) Characterization of two genes (hup D and hupE) required for hydrogenase activity in Azotobacter chroococcum. FEMS Microbiol Lett 96: 93–102
Eberz G, Friedrich B (1991) Three trans-acting regulatory functions control hydrogenase synthesis in Alcaligenes eutrophus. J Bacteriol 173: 1845–1854
Eisenberg D, Schwarz E, Komaromy M, Wall R (1984) Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol 179: 125–142
Eitinger T, Friedrich B (1991) Cloning, nucleotide sequence, and heterologous expression of a high-affinity nickel transport gene from Alcaligenes eutrophus. J Biol Chem 266: 3222–3227
Friedrich CG, Friedrich B, Bowien B (1981) Formation of enzymes of autotrophic metabolism during heterotrophic growth of Alcaligenes eutrophus. J Gen Microbiol 122: 69–78
Friedrich CG, Schneider K, Friedrich B (1982) Nickel in the catalytically active hydrogenase of Alcaligenes eutrophus. J Bacteriol 152: 42–48
Henikoff S (1984) Unidirectional digestion with exonuclease III creates targeted break points for DNA sequencing. Gene 28: 351–359
Hidalgo E, Palacios JM, Murillo J, Ruiz-Argüeso T (1992) Nucleotide sequence and characterization of four additional genes of the hydrogenase structural operon from Rhizobium leguminosarum bv. viciae. J Bacteriol 174: 4130–4139
Jacobi A, Rossmann R, Böck A (1992) The hyp operon gene products are required for the maturation of catalytically active hydrogenase isoenzymes in Escherichia coli. Arch Microbiol 158: 444–451
Jones BD, Mobley HLT (1989) Proteus mirabilis urease: nucleotide sequence determination and comparison with jack bean urease. J Bacteriol 171: 6414–6422
Kortlüke C, Friedrich B (1992) Maturation of membrane-bound hydrogenase of Alcaligenes eutrophus H16. J Bacteriol 174: 6290–6293
Kortlüke C, Hogrefe C, Eberz G, Pühler A, Friedrich B (1987) Genes of lithoautotrophic metabolism are clustered on the megaplasmid pHG1 in Alcaligenes eutrophus. Mol Gen Genet 210: 122–128
Kortlüke C, Horstmann K, Schwartz E, Rohde M, Binsack R, Friedrich B (1992) A gene complex coding for the membranebound hydrogenase of Alcaligenes eutrophus H16. J Bacteriol 174: 6277–6289
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–132
Lee MH, Mulrooney SB, Renner MJ, Markowicz Y, Hausinger RP (1992) Klebsiella aerogenes urease gene cluster: sequence of ure D and demonstration that four accessory genes (ureD, ureE, ureF, and ureG) are involved in nickel metallocenter biosynthesis. J Bacteriol 174: 4324–4330
Lutz S, Jacobi A, Schlensog V, Böhm R, Sawers G, Böck A (1991) Molecular characterization of an operon (hyp) necessary for the activity of the three hydrogenase isoenzymes in Escherichia coli. Mol Microbiol 5: 123–135
Maier T, Jacobi A, Sauter M, Lottspeich F, Böck A (1993) The product of the hypB gene which is required for nickel incorporation into hydrogenases in a novel G protein. J. Bacteriol 175: 630–635
Mulrooney SB, Hausinger RP (1990) Sequence of the Klebsiella aerogenes urease genes and evidence for accessory proteins facilitating nickel incorporation. J Bacteriol 172: 5837–5843
Normark S, Bergström S, Edlund T, Grundström G, Jaurin B, Lindberg FP, Olsson O (1983) Overlapping genes. Annu Rev Genet 17: 499–525
Przybyla AE, Robbins J, Menon N, Peck HD (1992) Structurefunction relationships among the nickel-containing hydrogenases. FEMS Microbiol Rev 88: 109–136
Rey L, Murillo J, Hernando Y, Hidalgo E, Cabrera E, Imperial J, Ruiz-Argüeso T (1993) Molecular analysis of a microaerobically induced operon required for hydrogenase synthesis in Rhizobium leguminosarum bv. viciae. Mol Microbiol (in press)
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Sanger F, Nicklen S, Coulsen AR (1977) DNA-sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74: 5463–5467
Schlegel HG, Kaltwasser H, Gottschalk G (1961) Ein Submersverfahren zur Kultur wasserstoffoxidierender Bakterien: wachstumsphysiologische Untersuchungen. Arch Mikrobiol 38: 209–222
Schneider K, Schlegel HG (1976) Purification and properties of soluble hydrogenase from Alcaligenes eutrophus H16. Biochim Biophys Acta 452: 66–80
Tran-Betcke A, Warnecke U, Böcker C, Zaborosch C, Friedrich B (1990) Cloning and nucleotide sequences of the genes for the subunits of NAD-reducing hydrogenase of Alcaligenes eutrophus H16. J Bacteriol 172: 2920–2929
Tabor S, Richardson CC (1985) A bacteriophage T7 polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA 82: 1074–1078
Xu H-W, Wall JD (1991) Clustering of genes necessary for hydrogen oxidation in Rhodobacter capsulatus. J Bacteriol 173: 2401–2405
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Dernedde, J., Eitinger, M. & Friedrich, B. Analysis of a pleiotropic gene region involved in formation of catalytically active hydrogenases in Alcaligenes eutrophus H16. Arch. Microbiol. 159, 545–553 (1993). https://doi.org/10.1007/BF00249034
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/BF00249034