Abstract
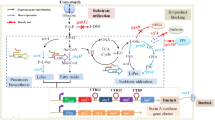
Signal peptide (SP) is an important factor and biobrick in the production and secretion of recombinant proteins. The aim of this study was in silico and in vivo analysis of SPs effect on the production of recombinant glucose oxidase (GOX) in Yarrowia lipolytica. Several in silico softwares, namely SignalP4, Signal-CF, Phobius, WolfPsort 0.2, SOLpro and ProtParam, were used to analyse the potential of 15 endogenous and exogenous SPs for the secretion of recombinant GOX in Y. lipolytica. According to in silico results, the SP of GOX was predicted as suitable in terms of high secretory potential and of protein solubility and stability which is chosen for in vivo analysis. The recombinant Y. lipolytica strain produced 280 U/L of extracellular GOX after 7 days in YPD medium. The results show that the SP of GOX can be applied to efficient production of extracellular heterologous proteins and metabolic engineering in Y. lipolytica.




Similar content being viewed by others
References
Bankar SB, Bule MV, Singhal RS, Ananthanarayan L (2009) Glucose oxidase—an overview. Biotechnol Adv 27:489–501
Baradaran A, Sieo CC, Foo HL, Illias RM, Yusoff K, Rahim RA (2013) Cloning and in silico characterization of two signal peptides from Pediococcus pentosaceus and their function for the secretion of heterologous protein in Lactococcus lactis. Biotechnol Lett 35:233–238
Beckerich JM, Boisramé-Baudevin A, Gaillardin C (1998) Yarrowia lipolytica: a model organism for protein secretion studies. Int Microbiol 1:123–130
Cereghino GPL, Cereghino JL, Ilgen C, Cregg JM (2002) Production of recombinant proteins in fermenter cultures of the yeast Pichia pastoris. Curr Opin Biotechnol 13:329–332
Choi J, Lee S (2004) Secretory and extracellular production of recombinant proteins using Escherichia coli. Appl Microbiol Biotechnol 64:625–635
Chou KC (2002) Prediction of protein signal sequences. Curr Protein Pept Sci 3:615–622
Chou KC, Shen HB (2007) Signal-CF: a subsite-coupled and window-fusing approach for predicting signal peptides. Biochem Biophys Res Comm 357:633–640
Damasceno LM, Huang CJ, Batt CA (2012) Protein secretion in Pichia pastoris and advances in protein production. Appl Microbiol Biotechnol 93:31–39
Darvishi F (2012) Expression of native and mutant extracellular lipases from Yarrowia lipolytica in Saccharomyces cerevisiae. Microb Biotechnol 5:634–641
Darvishi F, Destain J, Nahvi I, Thonart P, Zarkesh-Esfahani H (2011) High-level production of extracellular lipase by Yarrowia lipolytica mutants from methyl oleate. N Biotechnol 28:756–760
Darvishi F, Destain J, Nahvi I, Thonart P, Zarkesh-Esfahani H (2012) Effect of additives on freeze-drying and storage of Yarrowia lipolytica lipase. Appl Biochem Biotechnol 168:1101–1107
Darvishi F, Fathi Z, Ariana M, Moradi H (2017) Yarrowia lipolytica as a workhorse for biofuel production. Biochem Eng J 127:87–96
Darvishi F, Ariana M, Marella ER, Borodina I (2018) Advances in synthetic biology of oleaginous yeast Yarrowia lipolytica for producing non-native chemicals. Appl Microbiol Biotechnol 102:5925–5938
Darvishi Harzevili F (2014) Biotechnological applications of the yeast Yarrowia lipolytica. Springer, Berlin
Emanuelsson O, Nielsen H, Brunak S, Von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300:1005–1016
Frederick KR, Tung J, Emerick RS, Masiarz FR, Chamberlain SH, Vasavada A, Rosenberg S, Chakraborty S, Schopfer L, Schopter L (1990) Glucose oxidase from Aspergillus niger. cloning, gene sequence, secretion from Saccharomyces cerevisiae and kinetic analysis of a yeast-derived enzyme. J Biol Chem 265:3793–3802
Gasteiger E, Hoogland C, Gattiker A, Duvaud SE, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. Springer, Berlin
Ghasemi Y, Dabbagh F, Rasoul-Amini S, Haghighi AB, Morowvat MH (2012) The possible role of HSPs on Behçet’s disease: a bioinformatic approach. Comput Biol Med 42:1079–1085
Gray GL, Baldridge JS, McKeown KS, Heyneker HL, Chang CN (1985) Periplasmic production of correctly processed human growth hormone in Escherichia coli: natural and bacterial signal sequences are interchangeable. Gene 39:247–254
Gu L, Zhang J, Du G, Chen J (2015) Multivariate modular engineering of the protein secretory pathway for production of heterologous glucose oxidase in Pichia pastoris. Enzym Microb Tech 68:33–42
Hatzinikolaou D, Hansen O, Macris B, Tingey A, Kekos D, Goodenough P, Stougaard P (1996) A new glucose oxidase from Aspergillus niger: characterization and regulation studies of enzyme and gene. Appl Microbiol Biotechnol 46:371–381
Hodgkins M, Sudbery P, Mead D, Balance DJ, Goodey A (1993) Expression of the glucose oxidase gene from Aspergillus niger in Hansenula polymorpha and its use as a reporter gene to isolate regulatory mutations. Yeast 9:625–635
Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier C, Nakai K (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35:585–587
Humphreys DP, Sehdev M, Chapman AP, Ganesh R, Smith BJ, King LM, Glover DJ, Reeks DG, Stephens PE (2000) High-level periplasmic expression in Escherichia coli using a eukaryotic signal peptide: importance of codon usage at the 5′ end of the coding sequence. Protein Expr Purif 20:252–264
Käll L, Krogh A, Sonnhammer EL (2004) A combined transmembrane topology and signal peptide prediction method. J Mol Biol 338:1027–1036
Kapat A, Jung JK, Park YH (1998) Improvement of extracellular recombinant glucose oxidase production in fed-batch culture of Saccharomyces cerevisiae: effect of different feeding strategies. Biotechnol Lett 20:319–323
Khadivi Derakshan F, Darvishi F, Dezfulian M, Madzak C (2017) Expression and characterization of glucose oxidase from Aspergillus niger in Yarrowia lipolytica. Mol Biotechnol 59:307–314
Kleppe K (1966) The effect of hydrogen peroxide on glucose oxidase from Aspergillus niger. Biochemistry 5:139–143
Kober L, Zehe C, Bode J (2013) Optimized signal peptides for the development of high expressing CHO cell lines. Biotechnol Bioeng 110:1164–1173
Liang S, Li C, Ye Y, Lin Y (2013) Endogenous signal peptides efficiently mediate the secretion of recombinant proteins in Pichia pastoris. Biotechnol Lett 35:97–105
Liu H, Yang J, Ling JG, Chou KC (2005) Prediction of protein signal sequences and their cleavage sites by statistical rulers. Biochem Biophys Res Comm 338:1005–1011
Macauley-Patrick S, Fazenda ML, McNeil B, Harvey LM (2005) Heterologous protein production using the Pichia pastoris expression system. Yeast 22:249–270
Madzak C (2015) Yarrowia lipolytica: recent achievements in heterologous protein expression and pathway engineering. Appl Microbiol Biotechnol 99:4559–4577
Madzak C, Beckerich JM (2013) Heterologous protein expression and secretion in Yarrowia lipolytica. In: Barth G (ed) Yarrowia lipolytica: biotechnological applications. Springer, Berlin
Madzak C, Gaillardin C, Beckerich JM (2004) Heterologous protein expression and secretion in the non-conventional yeast Yarrowia lipolytica: a review. J Biotechnol 109:63–81
Madzak C, Nicaud JM, Gaillardin C (2005a) Yarrowia lipolytica. Production of recombinant proteins: novel microbial and eukaryotic expression systems. Springer, Berlin
Madzak C, Otterbein L, Chamkha M, Moukha S, Asther M, Gaillardin C, Beckerich JM (2005b) Heterologous production of a laccase from the basidiomycete Pycnoporus cinnabarinus in the dimorphic yeast Yarrowia lipolytica. FEMS Yeast Res 5:635–646
Magnan CN, Randall A, Baldi P (2009) SOLpro: accurate sequence-based prediction of protein solubility. Bioinformatics 25:2200–2207
Makrides SC (1996) Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol Rev 60:512–538
Malherbe D, Du Toit M, Otero R, Van Rensburg P, Pretorius I (2003) Expression of the Aspergillus niger glucose oxidase gene in Saccharomyces cerevisiae and its potential applications in wine production. Appl Microbiol Biotechnol 61:502–511
Massahi A, Çalık P (2015) In-silico determination of Pichia pastoris signal peptides for extracellular recombinant protein production. J Theor Biol 364:179–188
Meng Y, Zhao M, Yang M, Zhang Q, Hao J (2014) Production and characterization of recombinant glucose oxidase from Aspergillus niger expressed in Pichia pastoris. Lett Appl Microbiol 58:393–400
Mirbagheri M, Nahvi I, Emtiazi G, Darvishi F (2011) Enhanced production of citric acid in Yarrowia lipolytica by Triton X-100. Appl Biochem Biotechnol 165:1068–1074
Mirbagheri M, Nahvi I, Emtiazi G, Mafakher L, Darvishi F (2012) Taxonomic characterization and potential biotechnological applications of Yarrowia lipolytica isolated from meat and meat products. Jundishapur J Microbiol 5:346–351
Möller EM, Bahnweg G, Sandermann H, Geiger HH (1992) A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res 20:6115–6116
Nezafat N, Ghasemi Y, Javadi G, Khoshnoud MJ, Omidinia E (2014) A novel multi-epitope peptide vaccine against cancer: an in silico approach. J Theor Biol 349:121–134
Nicaud JM, Madzak C, Van den broek P, Gysler C, Duboc P, Niederberger P, Gaillardin C (2002) Protein expression and secretion in the yeast Yarrowia lipolytica. FEMS Yeast Res 2:371–379
Nielsen H, Engelbrecht J, Brunak S, Von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng Des Sel 10:1–6
Park EH, Shin YM, Lim YY, Kwon TH, Kim DH, Yang MS (2000) Expression of glucose oxidase by using recombinant yeast. J Biotechnol 81:35–44
Petersen TN, Brunak S, Von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786
Rocha SN, Abrahão-Neto J, Cerdán ME, González-Siso MI, Gombert AK (2010) Heterologous expression of glucose oxidase in the yeast Kluyveromyces marxianus. Microb Cell Fact 9:1–4
Sagemark J, Kraulis P, Weigelt J (2010) A software tool to accelerate design of protein constructs for recombinant expression. Protein Expr Purif 72:175–178
Sibirny A, Madzak C, Fickers P (2014) Genetic engineering of non-conventional yeast for the production of valuable compounds. In: Darvishi F, Chen H (eds) Microbial biotechnology: progress and trends. CRC Press, Boca Raton
Tuteja R (2005) Type I signal peptidase: an overview. Arch Biochem Biophys 441:107–111
Wang M, Yang J, Chou KC (2005) Using string kernel to predict signal peptide cleavage site based on subsite coupling model. Amino Acids 28:395–402
Wong CM, Wong KH, Chen XD (2008) Glucose oxidase: natural occurrence, function, properties and industrial applications. Appl Microbiol Biotechnol 78:927–938
Xuan JW, Fournier P, Gaillardin C (1988) Cloning of the LYS5 gene encoding saccharopine dehydrogenase from the yeast Yarrowia lipolytica by target integration. Curr Genet 14:15–21
Yamaguchi M, Tahara Y, Nakano A, Taniyama T (2007) Secretory and continuous expression of Aspergillus niger glucose oxidase gene in Pichia pastoris. Protein Expr Purif 55:273–278
Zamani M, Nezafat N, Negahdaripour M, Dabbagh F, Ghasemi Y (2015) In silico evaluation of different signal peptides for the secretory production of human growth hormone in Escherichia coli. Int J Pept Res Therapeut 21:261–268
Zimmermann R, Eyrisch S, Ahmad M, Helms V (2011) Protein translocation across the ER membrane. Biochim Biophys Acta Biomembr 1808:912–924
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Darvishi, F., Zarei, A. & Madzak, C. In silico and in vivo analysis of signal peptides effect on recombinant glucose oxidase production in nonconventional yeast Yarrowia lipolytica. World J Microbiol Biotechnol 34, 128 (2018). https://doi.org/10.1007/s11274-018-2512-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-018-2512-x