Abstract
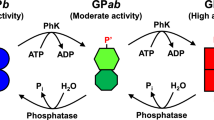
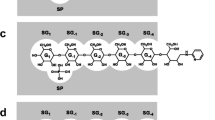
Glycogen phosphorylase (GP) is an allosteric enzyme whose catalytic site comprises six subsites (SG1, SG−1, SG−2, SG−3, SG−4, and SP) that are complementary to tandem five glucose residues and one inorganic phosphate molecule, respectively. In the catalysis of GP, the nonreducing-end glucose (Glc) of the maltooligosaccharide substrate binds to SG1 and is then phosphorolyzed to yield glucose 1-phosphate. In this study, we probed the catalytic site of rabbit muscle GP using pyridylaminated-maltohexaose (Glcα1–4Glcα1–4Glcα1–4Glcα1–4Glcα1–4GlcPA, where GlcPA = 1-deoxy-1-[(2-pyridyl)amino]-D-glucitol]; abbreviated as PA-0) and a series of specifically modified PA-0 derivatives (Glc m -AltNAc-Glc n -GlcPA, where m + n = 4 and AltNAc is 3-acetoamido-3-deoxy-D-altrose). PA-0 served as an efficient substrate for GP, whereas the other PA-0 derivatives were not as good as the PA-0, indicating that substrate recognition by all the SG1 –SG−4 subsites was important for the catalysis of GP. By comparing the initial reaction rate toward the PA-0 derivatives (V derivative) with that toward PA-0 (V PA-0), we found that the value of V derivative/V PA-0 decreased significantly as the level of allosteric activation of GP increased. These results suggest that some conformational changes have taken place in the maltooligosaccharide-binding region of the GP catalytic site during allosteric regulation.






Similar content being viewed by others
Abbreviations
- AltNAc:
-
3-acetoamido-3-deoxy-D-altrose
- CD:
-
Cyclodextrin
- CP-91149:
-
[R-(R*,S*)]-5-chloro-N-[3-(dimethylamino)-2-hydroxy-3-oxo-1-(phenylmethyl)propyl]-1H-indole-2-carboxamide
- DHB:
-
2,5-dihydroxybenzoic acid
- GDE:
-
Glycogen debranching enzyme
- Glc:
-
D-glucose
- Glc-1-P:
-
α-D-glucose 1-phosphate
- GlcPA:
-
1-deoxy-1-[(2-pyridyl)amino]-D-glucitol
- GP:
-
Glycogen phosphorylase
- HPLC:
-
High-performance liquid chromatography
- MALDI-TOF MS:
-
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
- MW:
-
Molecular weight
- α-NH2-CD:
-
3A-amino-3A-deoxy-(2AS,3AS)-α-cyclodextrin
- PA:
-
Pyridylamino
- Pi :
-
Inorganic phosphate
References
Sillerud, L.O., Shulman, R.G.: Structure and metabolism of mammalian liver glycogen monitored by carbon-13 nuclear magnetic resonance. Biochemistry. 22, 1087–1094 (1983)
Matsui, M., Kakuta, M., Misaki, A.: Comparison of the unit-chain distributions of glycogens from different biological sources, revealed by anion exchange chromatography. Biosci Biotechnol Biochem. 57, 623–627 (1993)
Roach, P.J., Depaoli-Roach, A.J., Hurley, T.D., Tagliabracci, V.S.: Glycogen and its metabolism: some new developments and old themes. Biochem J. 441, 763–787 (2012)
Titani, K., Koide, A., Hermann, J., Ericsson, L.H., Kumar, S., Wade, R.D., Walsh, K.A., Neurath, H., Fisher, E.H.: Complete amino acid sequence of rabbit muscle glycogen phosphorylase. Proc Natl Acad Sci U S A. 74, 4762–4766 (1977)
Tagaya, M., Fukui, T.: Catalytic reaction of glycogen phosphorylase reconstituted with a coenzyme-substrate conjugate. J Biol Chem. 259, 4860–4865 (1984)
Gordon, R.B., Brown, D.H., Brown, B.I.: Preparation and properties of the glycogen-debranching enzyme from rabbit liver. Biochim Biophys Acta. 289, 97–107 (1972)
Nakayama, A., Yamamoto, K., Tabata, S.: Identification of the catalytic residues of bifunctional glycogen debranching enzyme. J Biol Chem. 276, 28824–28828 (2001)
Sato, S., Ohi, T., Nishino, I., Sugie, H.: Confirmation of the efficiency of vitamin B6 supplementation for McArdle disease by follow-up muscle biopsy. Muscle Nerve. 45, 436–440 (2012)
Voet, D., Voet, J.D.: Biochemistry (third edition) pp. 626–656. John Wiley & sons Inc. Hoboken. (2004)
Berg, J.M., Tymoczko, J.L., Stryer, L.: Biochemistry (sixth edition) pp. 592–616. W. H. Freeman and company. N Y. (2007)
Miyagawa, D., Makino, Y., Sato, M.: Sensitive, nonradioactive assay of phosphorylase kinase through measurement of enhanced phosphorylase activity towards fluorogenic dextrin. J Biochem. 159, 239–246 (2016)
Madsen, N.B., Shechosky, S., Fletterick, R.J.: Site-site interactions in glycogen phosphorylase b probed by ligands specific for each site. Biochemistry. 22, 4460–4465 (1983)
Lowry, O.H., Schult, D.W., Passonneau, J.V.: Effects of adenylic acid on the kinetics of muscle phosphorylase a. J Biol Chem. 239, 1947–1953 (1964)
Rush, J.W.E., Spriet, L.L.: Skeletal muscle glycogen phosphorylase a kinetics: effects of adenine nucleotides and caffeine. J Appl Physiol. 91, 2071–2078 (2001)
Tanabe, S., Kobayashi, M., Matsuda, K.: Yeast glycogen phosphorylase: kinetic properties compared with muscle and potato enzymes. Agric Biol Chem. 52, 757–764 (1988)
Kasvinsky, P.J., Madsen, N.B., Fletterick, R.J., Sygusch, J.: X-ray crystallographic and kinetic studies of oligosaccharide binding to phosphorylase. J Biol Chem. 253, 1290–1296 (1978)
Sprang, S.R., Goldsmith, E.J., Fletterick, R.J., Withers, S.G., Madsen, N.B.: Catalytic site of glycogen phosphorylase: structure of the T state and specificity for α-D-glucose. Biochemistry. 21, 5364–5371 (1982)
Withers, S.G., Madsen, N.B., Sprang, S.R., Fletterick, R.J.: Catalytic site of glycogen phosphorylase: structural changes during activation and mechanistic implications. Biochemistry. 21, 5372–5382 (1982)
Hiromi, K.: Interpretation of dependency of rate parameters on the degree of polymerization of substrate in enzyme-catalyzed reactions. Evaluation of subsite affinities of exo-enzyme. Biochem Biophys Res Commun. 40, 1–6 (1970)
Hiromi, K., Nitta, Y., Numata, C., Ono, S.: Subsite affinities of glucoamylase: examination of the validity of the subsite theory. Biochim Biophys Acta. 302, 362–375 (1973)
Nitta, Y., Mizushima, M., Hiromi, K., Ono, S.: Influence of molecular structures of substrates and analogues on taka-amylase a catalyzed hydrolyses. I Effect of chain length of linear substrates J Biochem. 69, 567–576 (1971)
Konishi, Y., Kitazato, S., Nakatani, N.: Partial purification and characterization of acid and neutral α-glucosidases from preclimacteric banana pulp tissues. Biosci Biotechnol Biochem. 56, 2046–2051 (1992)
Fujita, K., Tahara, T., Koga, T., Imoto, T.: Enzymatic synthesis of specifically modified linear oligosaccharides from γ-cyclodextrin derivatives. Study on importance of acive sites of Taka amylase A. Bull Chem Soc Jpn. 62, 3150–5154 (1989)
Croft, A.P., Bartsch, R.A.: Synthesis of chemically modified cyclodextrins. Tetrahedron. 39, 1417–1474 (1983)
Walker, G.J., Whelan, W.J.: The mechanism of carbohydrase action: 8, structures of the muscle-phosphorylase limit dextrins of glycogen and amylopectin. Biochem J. 76, 264–268 (1960)
Hase, S., Ikenaka, T., Matsushima, Y.: Structure analyses of oligosaccharides by tagging of the reducing end sugars with a fluorescent compound. Biochem Biophys Res Commun. 85, 257–263 (1978)
Kuraya, N., Hase, S.: Release of O-linked sugar chains from glycoproteins with anhydrous hydrazine and pyridylamination of the sugar chains with improved reaction conditions. J Biochem. 112, 122–126 (1992)
Makino, Y., Omichi, K.: Acceptor specificity of 4-α-glucanotransferases of mammalian glycogen debranching enzymes. J Biochem. 139, 535–541 (2006)
Natsuka, S., Masuda, M., Sumiyoshi, W., Nakakita, S.: Improved method for drawing of a glycan map, and the first page of glycan atlas, which is a compilation of glycan maps for a whole organism. PLoS One. 9, e102219 (2014)
Makino, Y., Omichi, K., Hase, S.: Analysis of oligosaccharide structures from the reducing end terminal by combining partial acid hydrolysis and a two-dimensional sugar map. Anal Biochem. 264, 172–179 (1998)
Day, A.G., Parsonage, D., Ebel, S., Brown, T., Fersht, A.R.: Barnase has subsites that give rise to large rate enhancements. Biochemistry. 31, 6390–6395 (1992)
Buckle, A.M., Fersht, A.R.: Subsite binding in an RNase: structure of a barnase–tetranucleotide complex at 1.76-Å resolution. Biochemistry. 33, 1644–1653 (1994)
Burkhardt, G., Wegener, G.: Glycogen phosphorylase from flight muscle of the hawk moth Manduca sexta: purification and properties of three interconvertible forms and the effect of flight on their interconversion. J Comp Physiol B. 164, 261–271 (1994)
Makino, Y., Omichi, K.: Sensitive assay of glycogen phosphorylase activity by analysing the chain-lengthening action on a fluorogenic maltooligosaccharide derivative. J Biochem. 146, 71–76 (2009)
Lineweaver, H., Burk, D.: The determination of enzyme dissociation constants. J Am Chem Soc. 56, 658–666 (1934)
Makino, Y., Fujii, Y., Taniguchi, M.: Properties and functions of the storage sites of glycogen phosphorylase. J Biochem. 157, 451–458 (2015)
Barford, D., Johnson, L.N.: The allosteric transition of glycogen phosphorylase. Nature. 340, 609–616 (1989)
Buchbinder, J.L., Rath, V.L., Fletterick, R.J.: Structural relationships among regulated and unregulated phosphorylases. Annu Rev Biophys Biomol Struct. 30, 191–209 (2001)
Leonidas, D.D., Oikonomakos, N.G., Papageorgiou, A.C., Xenakis, A., Cazianis, C.T., Bem, F.: The ammonium sulfate activation of phosphorylase b. FEBS Lett. 261, 23–27 (1990)
Ishimizu, T., Hase, S.: Substrate recognition by sugar chain-related enzymes: recognition of a large area of substrates and its strictness and tolerance. Trends Glycosci Glycotechnol. 17, 215–227 (2005)
Okubo, M., Horinishi, A., Takeuchi, M., Suzuki, Y., Sakura, N., Hasegawa, Y., Igarashi, T., Goto, K., Tahara, H., Uchimoto, S., Omichi, K., Kanno, H., Hayasaka, K., Murase, T.: Heterogeneous mutations in the glycogen-debranching enzyme gene are responsible for glycogen storage disease type IIIa in Japan. Hum Genet. 106, 108–115 (2000)
Zhai, L., Feng, L., Xia, L., Yin, H., Xiang, S.: Crystal structure of glycogen debranching enzyme and insights into its catalysis and disease-causing mutations. Nat Commun. 7, 11229 (2016)
Watanabe, Y., Makino, Y., Omichi, K.: Donor substrate specificity of 4-α-glucanotransferase of porcine liver glycogen debranching enzyme and complementary action to glycogen phosphorylase on debranching. J Biochem. 143, 435–440 (2008)
Somsak, L., Czifrak, K., Toth, M., Bokor, E., Chrysina, E.D., Alexacou, K.M., Hayes, J.M., Tiraidis, C., Lazoura, E., Leonidas, D.D., Zographos, S.E., Oikonomakos, N.: New inhibitors of glycogen phosphorylase as potential antidiabetic agents. Curr Med Chem. 15, 2933–2983 (2008)
Martin, W.H., Hoover, D.J., Armento, S.J., Stock, I.A., McPherson, R.K., Danley, D.E., Stevenson, R.W., Barrett, E.J., Treadway, J.L.: Discovery of a human liver glycogen phosphorylase inhibitor that lowers blood glucose in vivo. Proc Natl Acad Sci U S A. 95, 1776–1781 (1998)
Lerin, C., Montell, E., Nolasco, T., Garcia-Rocha, M., Guinovart, J.J., Gomez-Foix, A.M.: Regulation of glycogen metabolism in cultured human muscles by the glycogen phosphorylase inhibitor CP-91149. Biochem J. 378, 1073–1077 (2004)
Oikonomakos, N.G., Chrysina, E.D., Kosmopoulou, M.N., Leonidas, D.D.: Crystal structure of rabbit muscle glycogen phosphorylase a in a complex with a potential hypoglycaemic drug at 2.0 Å resolution. Biochim Biophys Acta. 1647, 325–332 (2003)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Nakamura, M., Makino, Y., Takagi, C. et al. Probing the catalytic site of rabbit muscle glycogen phosphorylase using a series of specifically modified maltohexaose derivatives. Glycoconj J 34, 563–574 (2017). https://doi.org/10.1007/s10719-017-9776-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10719-017-9776-5