Abstract
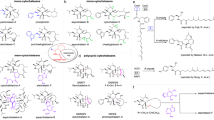
Fabclavines, unusual peptide–polyketide–polyamine hybrids, show broad-spectrum bioactivity against a variety of different organism like Gram-positive and -negative bacteria, fungi and protozoa. We elucidated the biosynthesis of these NRPS–PKS hybrids in Xenorhabdus szentirmaii by deletion of most genes encoded in the fabclavine BGC and subsequent analysis of produced fabclavine or polyamine intermediates. Thereby, we identified shortened fabclavines similar to the bioactive zeamines. Furthermore, we analyzed the thioester reductase FclG and the free-standing condensation domain-like protein FclL in detail and observed low substrate specificity for both enzymes.




Similar content being viewed by others
References
Bode HB (2009) Entomopathogenic bacteria as a source of secondary metabolites. Curr Opin Chem Biol 13:224–230. https://doi.org/10.1016/j.cbpa.2009.02.037
Bozhüyük KAJ, Fleischhacker F, Linck A, Wesche F, Tietze A, Niesert C-P, Bode HB (2018) De novo design and engineering of non-ribosomal peptide synthetases. Nat Chem 10:275–281. https://doi.org/10.1038/nchem.2890
Brachmann AO, Joyce SA, Jenke-Kodama H, Schwär G, Clarke DJ, Bode HB (2007) A type II polyketide synthase is responsible for anthraquinone biosynthesis in Photorhabdus luminescens. ChemBioChem 8:1721–1728. https://doi.org/10.1002/cbic.200700300
Cai X, Nowak S, Wesche F, Bischoff I, Kaiser M, Fürst R, Bode HB (2017) Entomopathogenic bacteria use multiple mechanisms for bioactive peptide library design. Nat Chem 9:379–386. https://doi.org/10.1038/nchem.2671
Chhabra A, Haque AS, Pal RK, Goyal A, Rai R, Joshi S, Panjikar S, Pasha S, Sankaranarayanan R, Gokhale RS (2012) Nonprocessive 2 + 2 e-off-loading reductase domains from mycobacterial nonribosomal peptide synthetases. Proc Natl Acad Sci USA 109:5681–5686. https://doi.org/10.1073/pnas.1118680109
Fuchs SW, Grundmann F, Kurz M, Kaiser M, Bode HB (2014) Fabclavines: bioactive peptide-polyketide-polyamino hybrids from Xenorhabdus. ChemBioChem 15:512–516. https://doi.org/10.1002/cbic.201300802
Keating TA, Marshall CG, Walsh CT (2000) Vibriobactin biosynthesis in Vibrio cholerae VibH is an amide synthase homologous to nonribosomal peptide synthetase condensation domains. Biochemistry 39:15513–15521. https://doi.org/10.1021/bi001651a
Kim I-H, Aryal SK, Aghai DT, Casanova-Torres ÁM, Hillman K, Kozuch MP, Mans EJ, Mauer TJ, Ogier J-C, Ensign JC, Gaudriault S, Goodman WG, Goodrich-Blair H, Dillman AR (2017) The insect pathogenic bacterium Xenorhabdus innexi has attenuated virulence in multiple insect model hosts yet encodes a potent mosquitocidal toxin. BMC Genom 18:927. https://doi.org/10.1186/s12864-017-4311-4
Kim I-H, Ensign J, Kim D-Y, Jung H-Y, Kim N-R, Choi B-H, Park S-M, Lan Q, Goodman WG (2017) Specificity and putative mode of action of a mosquito larvicidal toxin from the bacterium Xenorhabdus innexi. J Invertebr Pathol 149:21–28. https://doi.org/10.1016/j.jip.2017.07.002
Masschelein J, Clauwers C, Awodi UR, Stalmans K, Vermaelen W, Lescrinier E, Aertsen A, Michiels C, Challis GL, Lavigne R (2015) A combination of polyunsaturated fatty acid, nonribosomal peptide and polyketide biosynthetic machinery is used to assemble the zeamine antibiotics. Chem Sci 6:923–929. https://doi.org/10.1039/c4sc01927j
Masschelein J, Clauwers C, Stalmans K, Nuyts K, de Borggraeve W, Briers Y, Aertsen A, Michiels CW, Lavigne R (2015) The zeamine antibiotics affect the integrity of bacterial membranes. Appl Environ Microbiol 81:1139–1146. https://doi.org/10.1128/AEM.03146-14
McLennan AG (2006) The nudix hydrolase superfamily. Cell Mol Life Sci 63:123–143. https://doi.org/10.1007/s00018-005-5386-7
Müller S, Garcia-Gonzalez E, Mainz A, Hertlein G, Heid NC, Mösker E, van den Elst H, Overkleeft HS, Genersch E, Süssmuth RD (2014) Paenilamicin: structure and biosynthesis of a hybrid nonribosomal peptide/polyketide antibiotic from the bee pathogen Paenibacillus larvae. Angew Chem Int Ed Engl 53:10821–10825. https://doi.org/10.1002/anie.201404572
Reimer D, Bode HB (2014) A natural prodrug activation mechanism in the biosynthesis of nonribosomal peptides. Nat Prod Rep 31:154–159. https://doi.org/10.1039/c3np70081j
Strauch O, Ehlers R-U (1998) Food signal production of Photorhabdus luminescens inducing the recovery of entomopathogenic nematodes Heterorhabditis spp. in liquid culture. Appl Microbiol Biotechnol 50:369–374. https://doi.org/10.1007/s002530051306
Tobias NJ, Wolff H, Djahanschiri B, Grundmann F, Kronenwerth M, Shi Y-M, Simonyi S, Grün P, Shapiro-Ilan D, Pidot SJ, Stinear TP, Ebersberger I, Bode HB (2017) Natural product diversity associated with the nematode symbionts Photorhabdus and Xenorhabdus. Nat Microbiol 2:1676–1685. https://doi.org/10.1038/s41564-017-0039-9
Vingadassalon A, Lorieux F, Juguet M, Le Goff G, Gerbaud C, Pernodet J-L, Lautru S (2015) Natural combinatorial biosynthesis involving two clusters for the synthesis of three pyrrolamides in Streptomyces netropsis. ACS Chem Biol 10:601–610. https://doi.org/10.1021/cb500652n
Ziegenhorn J, Senn M, Bücher T (1976) Molar absorptivities of beta-NADH and beta-NADPH. Clin Chem 22:151–160
Acknowledgements
This work was supported by the DFG and the LOEWE Schwerpunkt MegaSyn supported by the State of Hessen. H.B.B. acknowledges the Deutsche Forschungsgemeinschaft for funding of the Impact II qTof mass spectrometer (INST 161/810-1). Furthermore, we would like to thank Dr. Hendrik Wolff and Peter Grün for technical assistance with the GC–MS measurements and Prof. Dr. Michael Karas for support with the MALDI-MS measurements.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Special Issue “Natural Product Discovery and Development in the Genomic Era 2019”.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wenski, S.L., Kolbert, D., Grammbitter, G.L.C. et al. Fabclavine biosynthesis in X. szentirmaii: shortened derivatives and characterization of the thioester reductase FclG and the condensation domain-like protein FclL. J Ind Microbiol Biotechnol 46, 565–572 (2019). https://doi.org/10.1007/s10295-018-02124-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-018-02124-8