Abstract
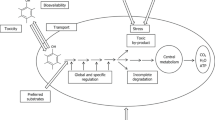
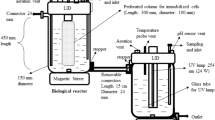
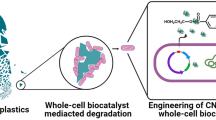
A microbial floc consisting of a community of microbes embedded in extracellular polymeric substances matrix can provide microbial resistances to toxic chemicals and harsh environments. Phenol is a toxic environmental pollutant and a typical lignin-derived phenolic inhibitor. In this study, we genetically engineered Escherichia coli cells by expressions of diguanylate cyclases (DGCs) to promote proteinaceous and aliphatic biofloc formation. Compared with the planktonic E. coli cells, the biofloc-forming cells improved phenol removal rate by up to 2.2-folds, due to their substantially improved tolerance (up to 149%) to phenol and slightly enhanced cellular activity (20%) of phenol hydroxylase (PheH). The engineered bioflocs also improved E. coli tolerance to other toxic compounds such as furfural, 5-hydroxymethylfurfural, and guaiacol. Additionally, the strategy of the engineered biofloc formation was applicable to Pseudomonas putida and enhanced its tolerance to phenol. This study highlights a strategy to form engineered bioflocs for improved cell tolerance and removal of toxic compounds, enabling their universality of use in bioproduction and bioremediation.






Similar content being viewed by others
References
Aparicio T, de Lorenzo V, Martinez-Garcia E (2018) CRISPR/Cas9-based Counterselection boosts recombineering efficiency in Pseudomonas putida. Biotechnol J 13(5):e1700161. https://doi.org/10.1002/biot.201700161
Arora PK, Bae H (2014) Bacterial degradation of chlorophenols and their derivatives. Microb Cell Factories 13(1):31. https://doi.org/10.1186/1475-2859-13-31
Beloin C, Roux A, Ghigo JM (2008) Escherichia coli biofilms. Curr Top Microbiol Immunol 322:249–289
Benedetti I, de Lorenzo V, Nikel PI (2016) Genetic programming of catalytic Pseudomonas putida biofilms for boosting biodegradation of haloalkanes. Metab Eng 33:109–118. https://doi.org/10.1016/j.ymben.2015.11.004
De N, Pirruccello M, Krasteva PV, Bae N, Raghavan RV, Sondermann H (2008) Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol 6(3):e67. https://doi.org/10.1371/journal.pbio.0060067
Engler C, Kandzia R, Marillonnet S (2008) A one pot, one step, precision cloning method with high throughput capability. PLoS One 3(11):e3647. https://doi.org/10.1371/journal.pone.0003647
Farhadian M, Duchez D, Vachelard C, Larroche C (2008) Monoaromatics removal from polluted water through bioreactors-a review. Water Res 42(6–7):1325–1341. https://doi.org/10.1016/j.watres.2007.10.021
Groning JA, Eulberg D, Tischler D, Kaschabek SR, Schlomann M (2014) Gene redundancy of two-component (chloro)phenol hydroxylases in Rhodococcus opacus 1CP. FEMS Microbiol Lett 361(1):68–75. https://doi.org/10.1111/1574-6968.12616
Gu H, Zhang J, Bao J (2015) High tolerance and physiological mechanism of Zymomonas mobilis to phenolic inhibitors in ethanol fermentation of corncob residue. Biotechnol Bioeng 112(9):1770–1782. https://doi.org/10.1002/bit.25603
Guvener ZT, Harwood CS (2007) Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol 66(6):1459–1473. https://doi.org/10.1111/j.1365-2958.2007.06008.x
Ha DG, O'Toole GA (2015) c-di-GMP and its effects on biofilm formation and dispersion: a Pseudomonas Aeruginosa review. Microbiol Spectr 3(2):MB-0003-2014. https://doi.org/10.1128/microbiolspec.MB-0003-2014
Hickman JW, Harwood CS (2008) Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol 69(2):376–389. https://doi.org/10.1111/j.1365-2958.2008.06281.x
Hickman JW, Tifrea DF, Harwood CS (2005) A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102(40):14422–14427. https://doi.org/10.1073/pnas.0507170102
Ibraheem O, Ndimba BK (2013) Molecular adaptation mechanisms employed by ethanologenic bacteria in response to lignocellulose-derived inhibitory compounds. Int J Biol Sci 9(6):598–612. https://doi.org/10.7150/ijbs.6091
Irankhah S, Abdi Ali A, Reza Soudi M, Gharavi S, Ayati B (2018) Highly efficient phenol degradation in a batch moving bed biofilm reactor: benefiting from biofilm-enhancing bacteria. World J Microbiol Biotechnol 34(11):164–113. https://doi.org/10.1007/s11274-018-2543-3
Jenal U, Reinders A, Lori C (2017) Cyclic di-GMP: second messenger extraordinaire. Nat Rev Microbiol 15(5):271–284. https://doi.org/10.1038/nrmicro.2016.190
Jones-Burrage SE, Kremer TA, McKinlay JB (2019) Cell aggregation and aerobic respiration are important for Zymomonas mobilis ZM4 survival in an aerobic minimal medium. Appl Environ Microbiol 85(10). https://doi.org/10.1128/AEM.00193-19
Jonsson LJ, Alriksson B, Nilvebrant NO (2013) Bioconversion of lignocellulose: inhibitors and detoxification. Biotechnol Biofuels 6(1):16. https://doi.org/10.1186/1754-6834-6-16
Kirchner U, Westphal AH, Muller R, van Berkel WJ (2003) Phenol hydroxylase from Bacillus thermoglucosidasius A7, a two-protein component monooxygenase with a dual role for FAD. J Biol Chem 278(48):47545–47553. https://doi.org/10.1074/jbc.M307397200
Klinke HB, Thomsen AB, Ahring BK (2004) Inhibition of ethanol-producing yeast and bacteria by degradation products produced during pre-treatment of biomass. Appl Microbiol Biotechnol 66(1):10–26. https://doi.org/10.1007/s00253-004-1642-2
Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S (2006) Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc Natl Acad Sci U S A 103(8):2839–2844. https://doi.org/10.1073/pnas.0511090103
Lee K, Yoon SS (2017) Pseudomonas aeruginosa biofilm, a programmed bacterial life for fitness. J Microbiol Biotechnol 27(6):1053–1064. https://doi.org/10.4014/jmb.1611.11056
Lee TS, Krupa RA, Zhang F, Hajimorad M, Holtz WJ, Prasad N, Lee SK, Keasling JD (2011) BglBrick vectors and datasheets: a synthetic biology platform for gene expression. J Biol Eng 5:12. https://doi.org/10.1186/1754-1611-5-12
Li Q, Zhao XQ, Chang AK, Zhang QM, Bai FW (2012) Ethanol-induced yeast flocculation directed by the promoter of TPS1 encoding trehalose-6-phosphate synthase 1 for efficient ethanol production. Metab Eng 14(1):1–8. https://doi.org/10.1016/j.ymben.2011.12.003
Morgan JL, McNamara JT, Zimmer J (2014) Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat Struct Mol Biol 21(5):489–496. https://doi.org/10.1038/nsmb.2803
Nicastro GG, Kaihami GH, Pereira TO, Meireles DA, Groleau MC, Deziel E, Baldini RL (2014) Cyclic-di-GMP levels affect Pseudomonas aeruginosa fitness in the presence of imipenem. Environ Microbiol 16(5):1321–1333. https://doi.org/10.1111/1462-2920.12422
Ojima Y, Azuma M, Taya M (2018) Inducing flocculation of non-floc-forming Escherichia coli cells. World J Microbiol Biotechnol 34(12):185–188. https://doi.org/10.1007/s11274-018-2563-z
Ojima Y, Nguyen MH, Yajima R, Taya M (2015) Flocculation of Escherichia coli cells in association with enhanced production of outer membrane vesicles. Appl Environ Microbiol 81(17):5900–5906. https://doi.org/10.1128/AEM.01011-15
Pontrelli S, Chiu TY, Lan EI, Chen FY, Chang P, Liao JC (2018) Escherichia coli as a host for metabolic engineering. Metab Eng. https://doi.org/10.1016/j.ymben.2018.04.008
Saa L, Jaureguibeitia A, Largo E, Llama MJ, Serra JL (2010) Cloning, purification and characterization of two components of phenol hydroxylase from Rhodococcus erythropolis UPV-1. Appl Microbiol Biotechnol 86(1):201–211. https://doi.org/10.1007/s00253-009-2251-x
Sharma G, Sharma S, Sharma P, Chandola D, Dang S, Gupta S, Gabrani R (2016) Escherichia coli biofilm: development and therapeutic strategies. J Appl Microbiol 121(2):309–319. https://doi.org/10.1111/jam.13078
Singh R, Paul D, Jain RK (2006) Biofilms: implications in bioremediation. Trends Microbiol 14(9):389–397. https://doi.org/10.1016/j.tim.2006.07.001
Spurbeck RR, Tarrien RJ, Mobley HL (2012) Enzymatically active and inactive phosphodiesterases and diguanylate cyclases are involved in regulation of motility or sessility in Escherichia coli CFT073. MBio 3(5). https://doi.org/10.1128/mBio.00307-12
Sureka K, Choi PH, Precit M, Delince M, Pensinger DA, Huynh TN, Jurado AR, Goo YA, Sadilek M, Iavarone AT, Sauer JD, Tong L, Woodward JJ (2014) The cyclic dinucleotide c-di-AMP is an allosteric regulator of metabolic enzyme function. Cell 158(6):1389–1401. https://doi.org/10.1016/j.cell.2014.07.046
Szokol J, Rucka L, Simcikova M, Halada P, Nesvera J, Patek M (2014) Induction and carbon catabolite repression of phenol degradation genes in Rhodococcus erythropolis and Rhodococcus jostii. Appl Microbiol Biotechnol 98(19):8267–8279. https://doi.org/10.1007/s00253-014-5881-6
Todhanakasem T, Yodsanga S, Sowatad A, Kanokratana P, Thanonkeo P, Champreda V (2018) Inhibition analysis of inhibitors derived from lignocellulose pretreatment on the metabolic activity of Zymomonas mobilis biofilm and planktonic cells and the proteomic responses. Biotechnol Bioeng 115(1):70–81. https://doi.org/10.1002/bit.26449
Tolker-Nielsen T (2015) Biofilm development. Microbiol Spectr 3(2):MB-0001-2014. https://doi.org/10.1128/microbiolspec.MB-0001-2014
Valentini M, Filloux A (2016) Biofilms and cyclic di-GMP (c-di-GMP) signaling: lessons from Pseudomonas aeruginosa and other Bacteria. J Biol Chem 291(24):12547–12555. https://doi.org/10.1074/jbc.R115.711507
Vedler E, Heinaru E, Jutkina J, Viggor S, Koressaar T, Remm M, Heinaru A (2013) Limnobacter spp. as newly detected phenol-degraders among Baltic Sea surface water bacteria characterised by comparative analysis of catabolic genes. Syst Appl Microbiol 36(8):525–532. https://doi.org/10.1016/j.syapm.2013.07.004
Wang B, Xu J, Gao J, Fu X, Han H, Li Z, Wang L, Tian Y, Peng R, Yao Q (2019) Construction of an Escherichia coli strain to degrade phenol completely with two modified metabolic modules. J Hazard Mater 373:29–38. https://doi.org/10.1016/j.jhazmat.2019.03.055
Wu Y, Ding Y, Cohen Y, Cao B (2015) Elevated level of the second messenger c-di-GMP in Comamonas testosteroni enhances biofilm formation and biofilm-based biodegradation of 3-chloroaniline. Appl Microbiol Biotechnol 99(4):1967–1976. https://doi.org/10.1007/s00253-014-6107-7
Xia J, Liu CG, Zhao XQ, Xiao Y, Xia XX, Bai FW (2018) Contribution of cellulose synthesis, formation of fibrils and their entanglement to the self-flocculation of Zymomonas mobilis. Biotechnol Bioeng 115(11):2714–2725. https://doi.org/10.1002/bit.26806
Xue C, Zhao XQ, Bai FW (2010) Effect of the size of yeast flocs and zinc supplementation on continuous ethanol fermentation performance and metabolic flux distribution under very high concentration conditions. Biotechnol Bioeng 105(5):935–944. https://doi.org/10.1002/bit.22610
Yoneda A, Henson WR, Goldner NK, Park KJ, Forsberg KJ, Kim SJ, Pesesky MW, Foston M, Dantas G, Moon TS (2016) Comparative transcriptomics elucidates adaptive phenol tolerance and utilization in lipid-accumulating Rhodococcus opacus PD630. Nucleic Acids Res 44(5):2240–2254. https://doi.org/10.1093/nar/gkw055
Zhao N, Bai Y, Liu CG, Zhao XQ, Xu JF, Bai FW (2014) Flocculating Zymomonas mobilis is a promising host to be engineered for fuel ethanol production from lignocellulosic biomass. Biotechnol J 9(3):362–371. https://doi.org/10.1002/biot.201300367
Acknowledgments
We thank Prof. Ningyi Zhou (Shanghai Jiaotong University, China) for his helpful suggestions and Prof. Luying Xun (Shandong University, China) and Prof. Rubing Liang (Shanghai Jiaotong University, China) for E. coli BL21(DE3)-groELS and P. putida KT2440, respectively.
Funding
This work was sponsored by the National Key R&D Program of China (2018YFA0901200), Science and Technology Commission of Shanghai Municipality (18JC1413600), and Natural Science Foundation of Shanghai (18ZR1420500). This work was also funded in part by Open Project Program of CAS Key Laboratory of Tropical Marine Bio-resources and Ecology (LMB), LMM and LAMB, SCSIO, CAS (Grant No. 2018011010).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 389 kb)
Rights and permissions
About this article
Cite this article
Jia, X., Zhang, S., Li, J. et al. Engineered bacterial biofloc formation enhancing phenol removal and cell tolerance. Appl Microbiol Biotechnol 104, 1187–1199 (2020). https://doi.org/10.1007/s00253-019-10289-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-10289-0