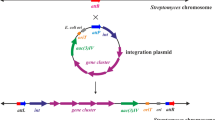
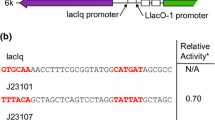
Abstract
Actinomycetes are the most important producers of secondary metabolites for medical, agricultural and industrial applications. Efficient engineering of bacterial genomes to improve their biosynthetic capabilities largely depends on the available arsenal of tools and vectors. One of the most widely used vector systems for actinomycetes is derived from the Streptomyces ghanaensis DSM2932 plasmid pSG5. pSG5 is a broad host range multicopy plasmid replicating via a rolling circle mechanism. The unique feature of pSG5, which distinguishes it from other Streptomyces plasmids, is its naturally thermosensitive mode of replication. This allows the efficient elimination of the plasmid from its host by simply shifting the incubation temperature to non-permissive 37–39 °C. This property makes pSG5 derivatives ideal facultative suicide vectors required for selection of gene disruption/gene replacement, transposon delivery or CRISPR/Cas9-mediated genome editing. Whereas these techniques depend on the fast elimination of the vector, stably replicating expression vectors for the production of recombinant proteins have been constructed more recently. This mini-review describes the generation and application of the pSG5 vector family, highlighting the specific features of the distinct vector plasmids.




Similar content being viewed by others
References
Arrowsmith TJ, Malpartida F, Sherman DH, Birch A, Hopwood DA, Robinson JA (1992) Characterisation of actI-homologous DNA encoding polyketide synthase genes from the monensin producer Streptomyces cinnamonensis. Mol Gen Genet 234:254–264
Baltz RH (2016) Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other actinomycetes. J Ind Microbiol Biotechnol 43:343–370
Behrmann I, Hillemann D, Pühler A, Strauch E, Wohlleben W (1990) Overexpression of a Streptomyces viridochromogenes gene (glnII) encoding a glutamine synthetase similar to those of eucaryotes confers resistance against the antibiotic phosphinothricyl-alanyl- ala. J Bacteriol 1729:5326–5334
Bibb MJ, Ward JM, Hopwood DA (1978) Transformation of plasmid DNA into Streptomyces at high frequency. Nature 274:398–400
Bierman M, Logan R, O’Brien K, Seno ET, Nagaraja-Rao R, Schoner BE (1992) Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43–49
Bilyk B, Weber S, Myronovskyi M, Bilyk O, Petzke L, Luzhetskyy A (2013) In vivo random mutagenesis of streptomycetes using mariner-based transposon Himar1. Appl Microbiol Biotechnol 97:351–359
Birch AW, Cullum J (1985) Temperature-sensitive mutants of the Streptomyces plasmid pIJ702. J Gen Microbiol 131:1299–1303
Blanco G, Pereda A, Mendez C, Salas JA (1992) Cloning and disruption of a fragment of Streptomyces halstedii DNA involved in the biosynthesis of a spore pigment. Gene 112:59–65
Chevillotte M, Menges R, Muth G, Wohlleben W, Stegmann E (2008) A quick and reliable method for monitoring gene expression in actinomycetes. J Biotechnol 135:262–265
Cormack BP, Valdivia RH, Falkow S (1996) FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33–38
Eichenlaub R (1979) Mutants of the mini-F plasmid pML31 thermosensitive in replication. J Bacteriol 138:559–566
Espinosa M, Cohen S, Couturier M, Del Solar G, Diaz Orejas R, Giraldo R, Janniere L, Miller C, Osborn M, Thomas CM (2000) Plasmid replication and copy number control. In: Thomas CM (ed) The horizontal gene pool. Harwood, Amsterdam, pp 1–48
Eys S, Schwartz D, Wohlleben W, Schinko E (2008) Three thioesterases are involved in the biosynthesis of phosphinothricin tripeptide in Streptomyces viridochromogenes Tu494. Antimicrob Agents Chemother 52:1686–1696
Fedoryshyn M, Petzke L, Welle E, Bechthold A, Luzhetskyy A (2008a) Marker removal from actinomycetes genome using Flp recombinase. Gene 419:43–47
Fedoryshyn M, Welle E, Bechthold A, Luzhetskyy A (2008b) Functional expression of the Cre recombinase in actinomycetes. Appl Microbiol Biotechnol 78:1065–1070
Feirtag JM, Petzel JP, Pasalodos E, Baldwin KA, McKay LL (1991) Thermosensitive plasmid replication, temperature-sensitive host growth, and chromosomal plasmid integration conferred by Lactococcus lactis subsp. cremoris lactose plasmids in Lactococcus lactis subsp. lactis. Appl Environ Microbiol 57:539–548
Fernandez-Martinez LT, Bibb MJ (2014) Use of the meganuclease I-SceI of Saccharomyces cerevisiae to select for gene deletions in actinomycetes. Sci Rep 4:7100. https://doi.org/10.1038/srep07100
Fink D, Falke D, Wohlleben W, Engels A (1999) Nitrogen metabolism in Streptomyces coelicolor A3(2): modification of glutamine synthetase I by an adenylyltransferase. Microbiology 145:2313–2322
Flett F, Mersinias V, Smith CP (1997) High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol Lett 155:223–229
Gentz R, Langner A, Chang AC, Cohen SN, Bujard H (1981) Cloning and analysis of strong promoters is made possible by the downstream placement of a RNA termination signal. Proc Natl Acad Sci U S A 78:4936–4940
Gerdes K, Ayora S, Canosa I, Ceglowski P, Diaz Orejas R, Franch T, Gultyaev AP, Bugge Jensen R, Kobayashi I, Macpherson C, Summers D, Thomas CM, Zielenkiewicz U (2000) Plasmid maintenance systems. In: Thomas CM (ed) The horizontal gene pool. Harwood, pp 49–86
Gust B, Challis GL, Fowler K, Kieser T, Chater KF (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci U S A 100:1541–1546
Herrmann S, Siegl T, Luzhetska M, Petzke L, Jilg C, Welle E, Erb A, Leadlay PF, Bechthold A, Luzhetskyy A (2012) Site-specific recombination strategies for engineering actinomycete genomes. Appl Environ Microbiol 78:1804–1812
Huang H, Zheng G, Jiang W, Hu H, Lu Y (2015) One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces. Acta Biochim Biophys Sin 47:231–243
Hulter N, Ilhan J, Wein T, Kadibalban AS, Hammerschmidt K, Dagan T (2017) An evolutionary perspective on plasmid lifestyle modes. Curr Opin Microbiol 38:74–80
Kataoka M, Kiyose YM, Michisuji Y, Horiguchi T, Seki T, Yoshida T (1994) Complete nucleotide sequence of the Streptomyces nigrifaciens plasmid, pSN22: genetic organization and correlation with genetic properties. Plasmid 32:55–69
Katz E, Thompson CJ, Hopwood DA (1983) Cloning and expression of the tyrosinase gene from Streptomyces antibioticus in Streptomyces lividans. J Gen Microbiol 129:2703–2714
Kieser T, Hopwood DA, Wright HM, Thompson CJ (1982) pIJ101, a multi-copy broad host-range Streptomyces plasmid: functional analysis and development of DNA cloning vectors. Mol Gen Genet 185:223–238
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces Genetics. The John Innes Foundation, Norwich
Kumagai T, Nakano T, Maruyama M, Mochizuki H, Sugiyama M (1999) Characterization of the bleomycin resistance determinant encoded on the transposon Tn5. FEBS Lett 442:34–38
Labes G, Simon R, Wohlleben W (1990) A rapid method for the analysis of plasmid content and copy number in various Streptomycetes grown on agar plates. Nucleic Acids Res 18:2197–2197
Labes G, Bibb M, Wohlleben W (1997) Isolation and characterization of a strong promoter element from the Streptomyces ghanaensis phage I19 using the gentamicin resistance gene (aacC1) of Tn1696 as reporter. Microbiol 143:1503–1512
Low ZJ, Pang LM, Ding Y, Cheang QW, Le Mai Hoang K, Thi Tran H, Li J, Liu XW, Kanagasundaram Y, Yang L, Liang ZX (2018) Identification of a biosynthetic gene cluster for the polyene macrolactam sceliphrolactam in a Streptomyces strain isolated from mangrove sediment. Sci Rep 8:1594. https://doi.org/10.1038/s41598-018-20018-8
Lussier FX, Denis F, Shareck F (2010) Adaptation of the highly productive T7 expression system to Streptomyces lividans. Appl Environ Microbiol 76:967–970
Maas RM, Gotz J, Wohlleben W, Muth G (1998) The conjugative plasmid pSG5 from Streptomyces ghanaensis DSM 2932 differs in its transfer functions from other Streptomyces rolling-circle-type plasmids. Microbiology 144:2809–2817
Maguin E, Duwat P, Hege T, Ehrlich D, Gruss A (1992) New thermosensitive plasmid for gram-positive bacteria. J Bacteriol 174:5633–5638
Martinez A, Kolvek SJ, Yip CL, Hopke J, Brown KA, MacNeil IA, Osburne MS (2004) Genetically modified bacterial strains and novel bacterial artificial chromosome shuttle vectors for constructing environmental libraries and detecting heterologous natural products in multiple expression hosts. Appl Environ Microbiol 70:2452–2463
Michta E, Schad K, Blin K, Ort-Winklbauer R, Rottig M, Kohlbacher O, Wohlleben W, Schinko E, Mast Y (2012) The bifunctional role of aconitase in Streptomyces viridochromogenes Tu494. Environ Microbiol 14:3203–3219
Million-Weaver S, Camps M (2014) Mechanisms of plasmid segregation: have multicopy plasmids been overlooked? Plasmid 75:27–36
Murakami T, Holt TG, Thompson CJ (1989) Thiostrepton-induced gene expression in Streptomyces lividans. J Bacteriol 171:1459–1466
Muth G, Wohlleben W, Puhler A (1988) The minimal replicon of the Streptomyces ghanaensis plasmid pSG5 identified by subcloning and Tn5 mutagenesis. Mol Gen Genet 211:424–429
Muth G, Nußbaumer B, Wohlleben W, Pühler A (1989) A vector system with temperature-sensitive replication for gene disruption and mutational cloning in streptomycetes. Mol Gen Genet 219:341–348
Muth G, Farr M, Hartmann V, Wohlleben W (1995) Streptomyces ghanaensis plasmid pSG5: nucleotide sequence analysis of the self-transmissible minimal replicon and characterization of the replication mode. Plasmid 33:113–126
Myronovskyi M, Welle E, Fedorenko V, Luzhetskyy A (2011) Beta-glucuronidase as a sensitive and versatile reporter in actinomycetes. Appl Environ Microbiol 77:5370–5383
Nakamura J, Kanno S, Kimura E, Matsui K, Nakamatsu T, Wachi M (2006) Temperature-sensitive cloning vector for Corynebacterium glutamicum. Plasmid 56(3):179–186
Olorunniji FJ, Rosser SJ, Stark WM (2016) Site-specific recombinases: molecular machines for the genetic revolution. Biochem J 473:673–684
Ostash B, Yushchuk O, Tistechok S, Mutenko H, Horbal L, Muryn A, Dacyuk Y, Kalinowski J, Luzhetskyy A, Fedorenko V (2015) The adpA-like regulatory gene from Actinoplanes teichomyceticus: in silico analysis and heterologous expression. World J Microbiol Biotechnol 31:1297–1301
Pernodet JL, Guerineau M (1981) Isolation and physical characterization of Streptomycete plasmids. Mol Gen Genet 182:53–59
Pernodet JL, Simonet JM, Guerineau M (1984) Plasmids in different strains of Streptomyces ambofaciens: free and integrated form of plasmid pSAM2. Mol Gen Genet 198:35–41
Petzke L, Luzhetskyy A (2009) In vivo Tn5-based transposon mutagenesis of Streptomycetes. Appl Microbiol Biotechnol 83:979–986
Pfeifer V, Nicholson GJ, Ries J, Recktenwald J, Schefer AB, Shawky RM, Schroder J, Wohlleben W, Pelzer S (2001) A polyketide synthase in glycopeptide biosynthesis: the biosynthesis of the non-proteinogenic amino acid (S)-3,5-dihydroxyphenylglycine. J Biol Chem 276:38370–38377
Pigac J, Vujaklija D, Toman Z, Gamulin V, Schrempf H (1988) Structural instability of a bifunctional plasmid pZG1 and single-stranded DNA formation in Streptomyces. Plasmid 19:222–230
Pridmore RD (1987) New and versatile cloning vectors with kanamycin-resistance marker. Gene 56:309–312
Qin Z, Munnoch JT, Devine R, Holmes NA, Seipke RF, Wilkinson KA, Wilkinson B, Hutchings MI (2017) Formicamycins, antibacterial polyketides produced by Streptomyces formicae isolated from African Tetraponera plant-ants. Chem Sci 8:3218–3227
Reuther J, Gekeler C, Tiffert Y, Wohlleben W, Muth G (2006a) Unique conjugation mechanism in mycelial streptomycetes: a DNA-binding ATPase translocates unprocessed plasmid DNA at the hyphal tip. Mol Microbiol 61:436–446
Reuther J, Wohlleben W, Muth G (2006b) Modular architecture of the conjugative plasmid pSVH1 from Streptomyces venezuelae. Plasmid 55:201–209
Rose K, Steinbuchel A (2002) Construction and intergeneric conjugative transfer of a pSG5-based cosmid vector from Escherichia coli to the polyisoprene rubber degrading strain Micromonospora aurantiaca W2b. FEMS Microbiol Lett 211:129–132
Sepulveda E, Vogelmann J, Muth G (2011) A septal chromosome segregator protein evolved into a conjugative DNA-translocator protein. Mob Genet Elements 1:225–229
Servín-González L, Sampieri A, Cabello J, Galván L, Juárez V, Castro C (1995) Sequence and functional analysis of the Streptomyces phaeochromogenes plasmid pJV1 reveals a modular organization of Streptomyces plasmids that replicate by rolling circle. Microbiology 141:2499–2510
Shetty RP, Endy D, Knight TF Jr (2008) Engineering BioBrick vectors from BioBrick parts. J Biol Eng 2:5. https://doi.org/10.1186/1754-1611-2-5
Silva F, Queiroz JA, Domingues FC (2012) Evaluating metabolic stress and plasmid stability in plasmid DNA production by Escherichia coli. Biotechnol Adv 30:691–708
Simon R, Priefer U, Pühler A (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnol 1:784–791
Solenberg PJ, Baltz RH (1991) Transposition of Tn5096 and other IS493 derivatives in Streptomyces griseofuscus. J Bacteriol 173:1096–1104
Solenberg PJ, Baltz RH (1994) Hypertransposing derivatives of the streptomycete insertion sequence IS493. Gene 147(1):47–54
Suzuki I, Kataoka M, Yoshida T, Seki T (2004) Lagging strand replication of rolling-circle plasmids in Streptomyces lividans: an RNA polymerase-independent primer synthesis. Arch Microbiol 181:305–313
Takano E, White J, Thompson CJ, Bibb MJ (1995) Construction of thiostrepton-inducible, high-copy-number expression vectors for use in Streptomyces spp. Gene 166:133–137
Thoma L, Muth G (2015) The conjugative DNA-transfer apparatus of Streptomyces. Int J Med Microbiol 305:224–229
Thoma L, Sepulveda E, Latus A, Muth G (2014) The stability region of the Streptomyces lividans plasmid pIJ101 encodes a DNA-binding protein recognizing a highly conserved short palindromic sequence motif. Front Microbiol 5:499. https://doi.org/10.3389/fmicb.2014.00499
Thoma L, Dobrowinski H, Finger C, Guezguez J, Linke D, Sepulveda E, Muth G (2015) A multiprotein DNA translocation complex directs intramycelial plasmid spreading during Streptomyces conjugation. MBio 6:e02559–e02514. https://doi.org/10.1128/mBio.02559-14
Thoma L, Vollmer B, Muth G (2016) Fluorescence microscopy of Streptomyces conjugation suggests DNA-transfer at the lateral walls and reveals the spreading of the plasmid in the recipient mycelium. Environ Microbiol 18:598–608
Tong Y, Charusanti P, Zhang L, Weber T, Lee SY (2015) CRISPR-Cas9 based engineering of Actinomycetal genomes. ACS Synth Biol 4:1020–1029
Tong Y, Robertsen HL, Blin K, Weber T, Lee SY (2018) CRISPR-Cas9 toolkit for Actinomycete genome editing. Methods Mol Biol 1671:163–184
Top EM, Moenne-Loccoz Y, Pembroke T, Thomas CM (2000) Phenotypic traits conferred by plasmids. In: Thomas CM (ed) The horizontal gene pool. Harwood Academic Publishers, pp 249–286
Vara J, Lewandowska-Skarbek M, Wang YG, Donadio S, Hutchinson CR (1989) Cloning of genes governing the deoxysugar portion of the erythromycin biosynthesis pathway in Saccharopolyspora erythraea (Streptomyces erythreus). J Bacteriol 171:5872–5881
Vogelmann J, Ammelburg M, Finger C, Guezguez J, Linke D, Flotenmeyer M, Stierhof YD, Wohlleben W, Muth G (2011a) Conjugal plasmid transfer in Streptomyces resembles bacterial chromosome segregation by FtsK/SpoIIIE. EMBO J 30:2246–2254
Vogelmann J, Wohlleben W, Muth G (2011b) Streptomyces conjugative elements. In: Dyson P (ed) Streptomyces—molecular biology and biotechnology. Caister Academic Press, Norfolk, pp 27–42
Volff JN, Altenbuchner J (1997) High frequency transposition of the Tn5 derivative Tn5493 in Streptomyces lividans. Gene 194:81–86
Wang Y, Cobb RE, Zhao H (2016) High-efficiency genome editing of Streptomyces species by an engineered CRISPR/Cas system. Methods Enzymol 575:271–284
Wehmeier UF (1995) New multifunctional Escherichia coli-Streptomyces shuttle vectors allowing blue-white screening on XGal plates. Gene 165:149–150
Wohlleben W, Arnold W, Bissonnette L, Pelletier A, Tanguay A, Roy PH, Gamboa GC, Barry GF, Aubert E, Davies J, Kagan SA (1989) On the evolution of Tn21-like multiresistance transposons: sequence analysis of the gene (aacC1) for gentamicin acetyltransferase-3-I(AAC(3)-I), another member of the Tn21-based expression cassette. Mol Gen Genet 217:202–208
Wolf T, Gren T, Thieme E, Wibberg D, Zemke T, Puhler A, Kalinowski J (2016) Targeted genome editing in the rare actinomycete Actinoplanes sp. SE50/110 by using the CRISPR/Cas9 system. J Biotechnol 231:122–128
Xu Z, Wang Y, Chater KF, Ou HY, Xu HH, Deng Z, Tao M (2017) Large-scale transposition mutagenesis of Streptomyces coelicolor identifies hundreds of genes influencing antibiotic biosynthesis. Appl Environ Microbiol 83. https://doi.org/10.1128/AEM.02889-16
Zechner EL, de la Cruz F, Eisenbrandt R, Grahn AM, Koraimann G, Lanka E, Muth G, Pansegrau W, Thomas CM, Wilks M, Zatyka M (2000) Conjugative-DNA transfer processes. In: Thomas CM (ed) The horizontal gene pool. Harwood Academic Publishers, pp 87–175
Zhang MM, Wong FT, Wang Y, Luo S, Lim YH, Heng E, Yeo WL, Cobb RE, Enghiad B, Ang EL, Zhao H (2017) CRISPR-Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. Nat Chem Biol 13:607–609. https://doi.org/10.1038/nchembio.2341
Zukowski MM, Miller L (1986) Hyperproduction of an intracellular heterologous protein in a sacUh mutant of Bacillus subtilis. Gene 46:247–255
Acknowledgements
The author thanks J. Authenrieth, A. Franco, A. Latus and E. Sulz for assistance in the vector constructions.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (SFB766).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declares that he has no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals.
Rights and permissions
About this article
Cite this article
Muth, G. The pSG5-based thermosensitive vector family for genome editing and gene expression in actinomycetes. Appl Microbiol Biotechnol 102, 9067–9080 (2018). https://doi.org/10.1007/s00253-018-9334-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-9334-5