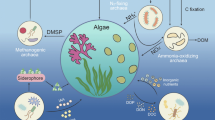
Abstract
Xestospongia muta is among the most emblematic sponge species inhabiting coral reefs of the Caribbean Sea. Besides being the largest sponge species growing in the Caribbean, it is also known to produce secondary metabolites. This study aimed to assess the effect of depth and season on the symbiotic bacterial dynamics and major metabolite profiles of specimens of X. muta thriving in a tropical marine biome (Portobelo Bay, Panamá), which allow us to determine whether variability patterns are similar to those reported for subtropical latitudes. The bacterial assemblages were characterized using Illumina deep-sequencing and metabolomic profiles using UHPLC-DAD-ELSD from five depths (ranging 9–28 m) across two seasons (spring and autumn). Diverse symbiotic communities, representing 24 phyla with a predominance of Proteobacteria and Chloroflexi, were found. Although several thousands of OTUs were determined, most of them belong to the rare biosphere and only 23 to a core community. There was a significant difference between the structure of the microbial communities in respect to season (autumn to spring), with a further significant difference between depths only in autumn. This was partially mirrored in the metabolome profile, where the overall metabolite composition did not differ between seasons, but a significant depth gradient was observed in autumn. At the phyla level, Cyanobacteria, Firmicutes, Actinobacteria, and Spirochaete showed a mild-moderate correlation with the metabolome profile. The metabolomic profiles were mainly characterized by known brominated polyunsaturated fatty acids. This work presents findings about the composition and dynamics of the microbial assemblages of X. muta expanding and confirming current knowledge about its remarkable diversity and geographic variability as observed in this tropical marine biome.





Similar content being viewed by others
References
Li CW, Chen JY, Hua TE (1998) Precambrian sponges with cellular structures. Science 279:879–882. https://doi.org/10.1126/science.279.5352.879
Yin Z, Zhu M, Davidson EH, Bottjer DJ, Zhao F, Tafforeau P (2015) Sponge grade body fossil with cellular resolution dating 60 Myr before the Cambrian. Proc Natl Acad Sci U S A 112:E1453–E1460. https://doi.org/10.1073/pnas.1414577112
Van-Soest R, Boury-Esnault N, Vacelet J et al (2012) Global diversity of sponges (Porifera). PLoS One 7:e35105. https://doi.org/10.1371/journal.pone.0035105
Gold DA, Grabenstatter J, De Mendoza A et al (2016) Sterol and genomic analyses validate the sponge biomarker hypothesis. Proc Natl Acad Sci 1–6:2684–2689. https://doi.org/10.1073/pnas.1512614113
Dayton PK (1989) Interdecadal variation in an antarctic sponge and its predators from oceanographic climate shifts. Science 245:1484–1486. https://doi.org/10.1126/science.245.4925.1484
Hentschel U, Piel J, Degnan SM, Taylor MW (2012) Genomic insights into the marine sponge microbiome. Nat Rev Microbiol 10:641–654. https://doi.org/10.1038/nrmicro2839
Bell JJ (2008) The functional roles of marine sponges. Estuar Coast Shelf Sci 79:341–353. https://doi.org/10.1016/j.ecss.2008.05.002
Deignan LK, Pawlik JR (2014) Perilous proximity: does the Janzen-Connell hypothesis explain the distribution of giant barrel sponges on a Florida coral reef? Coral Reefs 34:561–567. https://doi.org/10.1007/s00338-014-1255-x
Rix L, Naumann MS, de Goeij JM et al (2015) Coral mucus fuels the sponge loop in warm- and cold-water coral reef ecosystems. Scientific 6:1–11. https://doi.org/10.1038/srep18715
Pawlik JR, McMurray SE, Erwin P, Zea S (2015) No evidence for food limitation of Caribbean reef sponges: reply to Slattery & Lesser (2015). Mar Ecol Prog Ser 527:281–284. https://doi.org/10.3354/meps11308
De Goeij JM, Van Oevelen D, Vermeij MJA et al (2013) Surviving in a marine desert: the sponge loop retains resources within coral reefs. Science 342:108–110. https://doi.org/10.1126/science.1241981
Loh T-L, Pawlik JR (2014) Chemical defenses and resource trade-offs structure sponge communities on Caribbean coral reefs. Proc Natl Acad Sci U S A 111:4151–4156. https://doi.org/10.1073/pnas.1321626111
Zhang F, Blasiak LC, Karolin JO, Powell RJ, Geddes CD, Hill RT (2015) Phosphorus sequestration in the form of polyphosphate by microbial symbionts in marine sponges. Proc Natl Acad Sci 201423768:4381–4386. https://doi.org/10.1073/pnas.1423768112
Gatti S (2002) The role of sponges in high-Antarctic carbon and silicon cycling—a modelling approach. Ber Polarforsch Meeresforsch 434:1–134
Taylor MW, Radax R, Steger D, Wagner M (2007) Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev 71:295–347. https://doi.org/10.1128/MMBR.00040-06
Indraningrat AAG, Smidt H, Sipkema D (2016) Bioprospecting sponge-associated microbes for antimicrobial compounds. Mar Drugs 14:1–66. https://doi.org/10.3390/md14050087
Li Z-Y, He L-M, Wu J, Jiang Q (2006) Bacterial community diversity associated with four marine sponges from the South China Sea based on 16S rDNA-DGGE fingerprinting. J Exp Mar Biol Ecol 329:75–85. https://doi.org/10.1016/j.jembe.2005.08.014
Wang G (2006) Diversity and biotechnological potential of the sponge-associated microbial consortia. J Ind Microbiol Biotechnol 33:545–551. https://doi.org/10.1007/s10295-006-0123-2
Webster NS, Taylor MW (2012) Marine sponges and their microbial symbionts: love and other relationships. Environ Microbiol 14:335–346. https://doi.org/10.1111/j.1462-2920.2011.02460.x
Hentschel U, Usher KM, Taylor MW (2006) Marine sponges as microbial fermenters. FEMS Microbiol Ecol 55:167–177. https://doi.org/10.1111/j.1574-6941.2005.00046.x
Vacelet J, Donadey C (1975) Electron microscope study of the association between some sponges and bacteria. Bio Ecol 30:301–314
Webster NS, Thomas T (2016). Defining the Sponge Hologenome 7:1–14. https://doi.org/10.1128/mBio.00135-16.Invited
Ribes M, Jiménez E, Yahel G, López-Sendino P, Diez B, Massana R, Sharp JH, Coma R (2012) Functional convergence of microbes associated with temperate marine sponges. Environ Microbiol 14:1224–1239. https://doi.org/10.1111/j.1462-2920.2012.02701.x
Thoms C, Schupp P (2007) Chemical defense strategies in sponges: a review. Porifera Res Biodiversity Innov Sustain 627–637
Leal MC, Puga J, Serôdio J, Gomes NCM, Calado R (2012) Trends in the discovery of new marine natural products from invertebrates over the last two decades—where and what are we bioprospecting? PLoS One 7:e30580. https://doi.org/10.1371/journal.pone.0030580
Sacristán-Soriano O, Banaigs B, Casamayor EO, Becerro MA (2011) Exploring the links between natural products and bacterial assemblages in the sponge aplysina aerophoba. Appl Environ Microbiol 77:862–870. https://doi.org/10.1128/AEM.00100-10
Sipkema D, Franssen MCR, Osinga R, Tramper J, Wijffels RH (2005) Marine sponges as pharmacy. Mar Biotechnol 7:142–162. https://doi.org/10.1007/s10126-004-0405-5
Morinaka BI, Skepper CK, Molinski TF (2007) Ene-yne tetrahydrofurans from the sponge Xestospongia muta. Exploiting a Weak CD Effect for Assignment of Configuration 1:3–6
Schmitz FJ, Gopichand Y (1978) (7E,13E,15Z)-14,16-dibromo-7,13,15-hexadecatrien-5-ynoic acid. A novel dibromo acetylenic acid from the marine sponge Xestospongia muta. Tetrahedron Lett 3637–3640
Zhou X, Lu Y, Lin X, Yang B, Yang X, Liu Y (2011) Brominated aliphatic hydrocarbons and sterols from the sponge Xestospongia testudinaria with their bioactivities. Chem Phys Lipids 164:703–706. https://doi.org/10.1016/j.chemphyslip.2011.08.002
McMurray SE, Finelli CM, Pawlik JR (2015) Population dynamics of giant barrel sponges on Florida coral reefs. J Exp Mar Biol Ecol 473:73–80. https://doi.org/10.1016/j.jembe.2015.08.007
Gloeckner V, Wehrl M, Moitinho-Silva L, Gernert C, Schupp P, Pawlik JR, Lindquist NL, Erpenbeck D, Wörheide G, Hentschel U (2014) The HMA-LMA dichotomy revisited: an electron microscopical survey of 56 sponge species. Biol Bull 227:78–88
Zhou X, Xu T, Yang X-W, Huang R, Yang B, Tang L, Liu Y (2010) Chemical and biological aspects of marine sponges of the genus Xestospongia. Chem Biodivers 7:2201–2227. https://doi.org/10.1002/cbdv.201000024
Giannini A, Kushnir Y, Cane MA (2000) Interannual variability of Caribbean rainfall, ENSO, and the Atlantic Ocean. J Clim 13:297–311. https://doi.org/10.1175/1520-0442(2000)013<0297:IVOCRE>2.0.CO;2
McMurray SE, Blum JE, Pawlik JR (2008) Redwood of the reef: growth and age of the giant barrel sponge Xestospongia muta in the Florida keys. Mar Biol 155:159–171. https://doi.org/10.1007/s00227-008-1014-z
Griffiths R, Whiteley A (2000) Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA-and rRNA-based microbial community composition. Appl Environ Microbiol 66:1–5. https://doi.org/10.1128/AEM.66.12.5488-5491.2000.Updated
Simister RL, Schmitt S, Taylor MW (2011) Evaluating methods for the preservation and extraction of DNA and RNA for analysis of microbial communities in marine sponges. J Exp Mar Biol Ecol 397:38–43. https://doi.org/10.1016/j.jembe.2010.11.004
Bohorquez LC, Delgado-Serrano L, López G, Osorio-Forero C, Klepac-Ceraj V, Kolter R, Junca H, Baena S, Zambrano MM (2012) In-depth characterization via complementing culture-independent approaches of the microbial community in an acidic hot spring of the Colombian Andes. Microb Ecol 63:103–115. https://doi.org/10.1007/s00248-011-9943-3
Galvez E, Junca H, Riaño D (2012) Assessment of microbial composition and degradation functions in PAH contaminated neotropical Caribbean marine sediments (Cartagena Bay, Colombia). Universidad de Los Andes
Camarinha-Silva A, Juregui R, Chaves-Moreno D et al (2014) Comparing the anterior nare bacterial community of two discrete human populations using Illumina amplicon sequencing. Environ Microbiol 16:2939–2952. https://doi.org/10.1111/1462-2920.12362
Kuczynski J, Stombaugh J, Walters WA et al (2011) Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Bioinformatics Chapter 10:Unit 10.7. https://doi.org/10.1002/0471250953.bi1007s36
Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R (2010) PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. https://doi.org/10.1093/bioinformatics/btp636
Ivanišević J, Thomas OP, Lejeusne C, Chevaldonné P, Pérez T (2010) Metabolic fingerprinting as an indicator of biodiversity: towards understanding inter-specific relationships among Homoscleromorpha sponges. Metabolomics 7:289–304. https://doi.org/10.1007/s11306-010-0239-2
Greff S, Zubia M, Payri C, Thomas OP, Perez T (2017) Chemogeography of the red macroalgae Asparagopsis: metabolomics, bioactivity, and relation to invasiveness. Metabolomics 13:0. https://doi.org/10.1007/s11306-017-1169-z
Hamady M, Lozupone C, Knight R (2010) Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4:17–27. https://doi.org/10.1038/ismej.2009.97
Clarke KR, Warwick RM (2001) Change in marine communities: an approach to statistical analysis and interpretation, 2nd edition. Prim Plymouth UK p. 172
Anderson MJ (2001) A new method for non parametric multivariate analysis of variance. Austral Ecol 26:32–46. https://doi.org/10.1111/j.1442-9993.2001.01070.pp.x
McMurdie PJ, Holmes S (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. https://doi.org/10.1371/journal.pone.0061217
Hollister EB, Engledow AS, Hammett AJM, Provin TL, Wilkinson HH, Gentry TJ (2010) Shifts in microbial community structure along an ecological gradient of hypersaline soils and sediments. ISME J 4:829–838. https://doi.org/10.1038/ismej.2010.3
Barberan A, Bates ST, Casamayor EO, Fierer N (2012) Using network analysis to explore co-occurrence patterns in soil microbial communities. Isme J 6:343–351. https://doi.org/10.1038/ismej.2011.119
Fiore CL, Jarett JK, Lesser MP (2013) Symbiotic prokaryotic communities from different populations of the giant barrel sponge, Xestospongia muta. Microbiologyopen 2:938–952. https://doi.org/10.1002/mbo3.135
Montalvo NF, Hill RT (2011) Sponge-associated bacteria are strictly maintained in two closely related but geographically distant sponge hosts. Appl Environ Microbiol 77:7207–7216. https://doi.org/10.1128/AEM.05285-11
Thomas T, Moitinho-Silva L, Lurgi M, Björk JR, Easson C, Astudillo-García C, Olson JB, Erwin PM, López-Legentil S, Luter H, Chaves-Fonnegra A, Costa R, Schupp PJ, Steindler L, Erpenbeck D, Gilbert J, Knight R, Ackermann G, Victor Lopez J, Taylor MW, Thacker RW, Montoya JM, Hentschel U, Webster NS (2016) Diversity, structure and convergent evolution of the global sponge microbiome. Nat Commun 7:11870. https://doi.org/10.1038/ncomms11870
Webster NS, Taylor MW, Behnam F, Lücker S, Rattei T, Whalan S, Horn M, Wagner M (2010) Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ Microbiol 12:2070–2082. https://doi.org/10.1111/j.1462-2920.2009.02065.x
Schmitt S, Tsai P, Bell J, Fromont J, Ilan M, Lindquist N, Perez T, Rodrigo A, Schupp PJ, Vacelet J, Webster N, Hentschel U, Taylor MW (2012) Assessing the complex sponge microbiota: core, variable and species-specific bacterial communities in marine sponges. ISME J 6:564–576. https://doi.org/10.1038/ismej.2011.116
Simister RL, Deines P, Botté ES, Webster NS, Taylor MW (2012) Sponge-specific clusters revisited: a comprehensive phylogeny of sponge-associated microorganisms. Environ Microbiol 14:517–524. https://doi.org/10.1111/j.1462-2920.2011.02664.x
Patil A, Kokke W, Cochran S et al (1992) Brominated polyacetylenic acids from the marine sponge Xestospongia muta: inhibitors of HIV protease. J Nat Prod 55:1170–1177
Morrow KM, Fiore C, Lesser M (2016) Environmental drivers of microbial community shifts in the giant barrel sponge, Xestospongia muta, over a shallow to mesophotic depth gradient. Environ Microbiol n/a-n/a. https://doi.org/10.1111/1462-2920.13226
Olson JB, Gao X (2013) Characterizing the bacterial associates of three Caribbean sponges along a gradient from shallow to mesophotic depths. FEMS Microbiol Ecol 85:74–84. https://doi.org/10.1111/1574-6941.12099
Schmitt S, Angermeier H, Schiller R, Lindquist N, Hentschel U (2008) Molecular microbial diversity survey of sponge reproductive stages and mechanistic insights into vertical transmission of microbial symbionts. Appl Environ Microbiol 74:7694–7708. https://doi.org/10.1128/AEM.00878-08
Lesser MP, Fiore C, Slattery M, Zaneveld J (2016) Climate change stressors destabilize the microbiome of the Caribbean barrel sponge, Xestospongia muta. J Exp Mar Biol Ecol 475:11–18. https://doi.org/10.1016/j.jembe.2015.11.004
Wilkinson C (1978) Microbial associations in sponges. III. Ultrastructure of the in situ associations in coral reef sponges. Mar Biol 49:177–185
Burgsdorf I, Slaby BM, Handley KM, Haber M, Blom J, Marshall CW, Gilbert JA, Hentschel U, Steindler L (2015) Lifestyle evolution in cyanobacterial symbionts of sponges. MBio 6:1–14. https://doi.org/10.1128/mBio.00391-15
Usher KM (2008) The ecology and phylogeny of cyanobacterial symbionts in sponges. Mar Ecol 29:178–192. https://doi.org/10.1111/j.1439-0485.2008.00245.x
Freeman CJ, Thacker RW, Baker DM, Fogel ML (2013) Quality or quantity: is nutrient transfer driven more by symbiont identity and productivity than by symbiont abundance? ISME J 7:1116–1125. https://doi.org/10.1038/ismej.2013.7
Preston CM, Wu KY, Molinskit TF, Delong EF (1996) A psychrophilic crenarchaeon inhabits a marine sponge. Proc Natl Acad Sci U S A 93:6241–6246
Steger D, Ettinger-Epstein P, Whalan S, Hentschel U, de Nys R, Wagner M, Taylor MW (2008) Diversity and mode of transmission of ammonia-oxidizing archaea in marine sponges. Environ Microbiol 10:1087–1094. https://doi.org/10.1111/j.1462-2920.2007.01515.x
Zhang H, Sekiguchi Y, Hanada S et al (2003) Gemmatimonas aurantiaca gen. nov., sp. nov., a Gram-negative, aerobic, polyphosphate-accumulating micro-organism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov. Int J Syst Evol Microbiol 53:1155–1163. https://doi.org/10.1099/ijs.0.02520-0
Lesser MP (2006) Benthic–pelagic coupling on coral reefs: feeding and growth of Caribbean sponges. J Exp Mar Biol Ecol 328:277–288. https://doi.org/10.1016/j.jembe.2005.07.010
Unson MD, Holland ND, Faulkner DJ (1994) A brominated secondary metabolite synthesized by the cyanobacterial symbiont of a marine sponge and accumulation of the crystalline metabolite in the sponge tissue. Mar Biol 119:1–11. https://doi.org/10.1007/BF00350100
Kennedy J, Baker P, Piper C, Cotter PD, Walsh M, Mooij MJ, Bourke MB, Rea MC, O’Connor PM, Ross RP, Hill C, O’Gara F, Marchesi JR, Dobson ADW (2009) Isolation and analysis of bacteria with antimicrobial activities from the marine sponge Haliclona simulans collected from Irish waters. Mar Biotechnol 11:384–396. https://doi.org/10.1007/s10126-008-9154-1
Fiore CL, Labrie M, Jarett JK, Lesser MP (2015) Transcriptional activity of the giant barrel sponge, Xestospongia muta Holobiont: molecular evidence for metabolic interchange. Front. Microbiol. https://doi.org/10.3389/fmicb.2015.00364, 6
Acknowledgments
We thank the additional support from Dr. Regis Guillaume, Embassy of France in Colombia, and Raúl de León from Dive Adventure Panama and Portobelo Dive Center for his guidance and collaboration during the dive sampling, to Iris Plumeier and Silke Kahl for excellent technical assistance and to the members of the BIOMMAR research group for their helpful technical advices.
Funding
The authors would like to thank financial support granted by the European Commission to EC/FP7 research project consortium MAGICPAH (KBBE-2009-245226), to the Colombian Administrative Department for Science, Technology and Innovation - Colciencias for financial support through grants EcosNord-Colciencias (Convocatoria 652-2014) “International cooperation and research mobility grants in marine research between Colombia and France 2015-2017” and “Joven Investigador” 2012 fellowship to M. Villegas-Plazas and to Universidad de los Andes for the MSc scholarship to M. Villegas-Plazas.
Author information
Authors and Affiliations
Contributions
MV analyzed and interpreted the microbial community data and wrote the manuscript. OT analyzed and interpreted the metabolomic data. MO supported statistical analyses. HJ conceptualized, analyzed the results, and supervised the study. JS contributed with the experimental design and DP with the sequencing and analyzing the results. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable
Consent for Publication
Not applicable
Competing Interests
The authors declare that they have no competing interests.
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Villegas-Plazas, M., Wos-Oxley, M.L., Sanchez, J.A. et al. Variations in Microbial Diversity and Metabolite Profiles of the Tropical Marine Sponge Xestospongia muta with Season and Depth. Microb Ecol 78, 243–256 (2019). https://doi.org/10.1007/s00248-018-1285-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-018-1285-y