Abstract
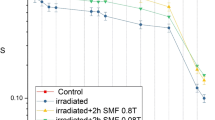
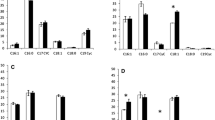
Several studies have investigated the effects of ionizing and non-ionizing radiations on microorganisms. However, the interaction between the magnetic field radiations and bacteria is less studied. The aim of our study was to study the effect of static magnetic field on the biofilm formation in Pseudomonas aeruginosa and its isogenic sod mutants. Our results revealed that the exposure to the static magnetic field (200 mT) increases significantly the swarming in the wild strain. The fliC gene expression did not show significant difference after 6 h exposure of the wild-type strain. The release of some compounds of the biofilm matrix such as rhamnolipids has been considerably enhanced after 6 h of exposure in the wild type. On the other hand, the pyocyanin and biofilm production was increased significantly in all strains compared to controls. Furthermore, our results revealed that the biofilm formation was confirmed by the pslA and ppyR gene expressions.





Similar content being viewed by others
References
Anderson GG, O’Toole GA (2008) Innate and induced resistance mechanisms of bacterial biofilms. Curr Top Microbiol Immunol 322:85–105
Arai H (2011) Regulation and function of versatile aerobic and anaerobic respiratory metabolism in Pseudomonas aeruginosa. Front Microbiol 2:103
Bellingham NF, Morgan JAW, Saunders JR, Winstanlley C (2001) Flagellin gene sequence variation in the genus Pseudomonas. Syst Appl Microbiol 24:157–165
Borlee BR, Goldman A, Murakami K, Samudrala R, Wozniak DJ, Parsek MR (2010) Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol 75:827–842
Branda SS, Vik S, Friedman L, Kolter R (2005) Biofilms: the matrix revisited. Trends Microbiol 13:20–26
Caiazza NC, Shanks RM, O'Toole GA (2005) Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J Bacteriol 187:7351–7361
Ceri H, Olson ME, Turner RJ (2010) Needed, new paradigms in antibiotic development. Expert Opin Pharmacother 11:1233–1237
Chandrasekaran EV, Bemiller JN (1980) Constituent analysis of glycosaminoglycans. In: Whistler RL, Wolfrom ML (eds) Methods in carbohydrate chemistry. Academic Press, New York, pp 89–96
Davey ME, Caiazza NC, O’Toole GA (2003) Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J Bacteriol 185:1027–1036
Dekimpe V, Déziel E (2009) Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR-specific factors. Microbiology 155(3):712–723
Déziel E, Lépine F, Milot S, Villemur R (2003) rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149:2005–2013
Dos Santos LO, Gonzales TA, Úbeda BT, Alegre RM (2012) Glutathione production using magnetic fields generated by magnets. Braz Arch Biol Technol 55:921–926
El May A, Snoussi S, Ben Miloud N, Maatouk I, Abdelmelek H, Ben Aissa R, Landoulsi A (2009) Effects of static magnetic fieldon cell growth, viability and differential gene expression in Salmonella. Foodborn Pathog Dis 6:547–552
Essar DW, Eberly L, Hadero A, Crawford IP (1990) Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol 172:884–900
Favre-Bonte S, Kohler T, Van Delden C (2003) Biofilm formation by Pseudomonas aeruginosa: role of the C4-AHL cell-to-cell signal and inhibition by azithromycin. J Antimicrob Chemother 52:598–604
Filipic J, Kraigher B, Tepus B, Kokol V, Mandic-Mulec I (2012) Effect of Low-density magnetic fields on the growth and activities of wastewater bacteria Escherichia coli and Pseudomonas putida. Bioresour Technol 120:225–232
Friedman L, Kolter R (2004) Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J Bacteriol 186:4457–4465
Ghadaksaz A, Fooladi AAI, Hosseini HM, Amin M (2014) The prevalence of some Pseudomonas virulence genes related to biofilm formation and alginate production among clinical isolates. J Appl Biomed 13:61–68
Grant CCR, Konkel ME, Cieplak JW, Tompkins LS (1993) Role of flagella in adherence, internalization, and translocation of Campylobacter jejuni in nonpolarized and polarized epithelial cell cultures. Infect Immun 30:1764–1771
Hall-Stoodley L, Stoodley P (2009) Evolving concepts in biofilm infections. Cell Microbiol 11:1034–1043
Hanini R, Chatti A, Ghorbel SB, Landoulsi A (2017) Role of sod gene in response to static magnetic fields in Pseudomonas aeruginosa. Curr Microbiol 74(8):930–937
Iiyama K, Chieda Y, Lee JM, Kusakabe T, Yasunaga-Aoki C, Shimizu S (2007) Effect of superoxide dismutase gene inactivation on virulence of Pseudomonas aeruginosa PAO1 toward the silkworm Bombyx mori. Appl Environ Microbiol 73(5):1569–1575
Intarak N, Muangsombut V, Vattanaviboon P, Stevens MP, Korbsrisate S (2014) Growth, motility and resistance to oxidative stress of the melioidosis pathogen Burkholderia pseudomallei are enhanced by epinephrine. Pathog Dis 72(1):24–31
Jackson KD, Starkey M, Kremer S, Parsek MR, Wozniak DJ (2004) Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J Bacteriol 186:4466–4475
Kim E-J, Sabra W, Zeng A-P (2003) Iron deficiency leads to inhibition of oxygen transfer and enhanced formation of virulence factors in cultures of Pseudomonas aeruginosa PAO1. Microbiology 149:2627–2634
Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jørgensen A, Mølin S, Tolker-Nielsen T (2003) Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol 48:1511–1524
Köhler T, Curty LK, Barja F, Van Delden C, Pechére JC (2000) Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J Bacteriol 182:5990–5996
Kurachi M (1958) Studies on the biosynthesis of pyocyanine. (II): isolation and determination of pyocyanine. Bull Inst Chem Res Kyoto Univ 36:174–187
Ma L, Conover M, Lu H, Parsek MR, Bayles K, Wozniak DJ (2009) Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog 5:e1000354
Matsukawa M, Greenberg EP (2004) Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J Bacteriol 186:4449–4456
Montie TC, Doyle-Huntzinger D, Craven RC, Holder IA (1982) Loss of virulence associated with absence of flagellum in an isogenic mutant of Pseudomonas aeruginosa in the burned-mouse model. Infect Immun 38:1296–1298
Murray TS, Kazmierczak BI (2006) FlhF is required for swimming and swarming in Pseudomonas aeruginosa. J Bacteriol 188:6995–7004
Nadell CD, Drescher K, Wingreen NS, Bassler BL (2015) Extracellular matrix structure governs invasion resistance in bacterial biofilms. ISME J 9:1700–1709
O'Toole GA, Kolter R (1998) Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304
Pacheco GJ, Reis RS, Fernandes ACLB, da Rocha SLG, Pereira MD, Perales J, Freire DMG (2012) Rhamnolipid production: effect of oxidative stress on virulence factors and proteome of Pseudomonas aeruginosa PA1. Appl Microbiol Biotechnol 95:1519–1529
Pearson JP, Van Delden C, Iglewski BH (1999) Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J Bacteriol 181:1203–1210
Potera C (1999) Forging a link between biofilm and disease. Science 283:1837–1839
Ramos I, Dietrich LEP, Price-Whelan A, Newman DK (2010) Phenazines affect biofilm formation by Pseudomonas aeruginosa in similar ways at various scales. Res Microbiol 161(3):187–191
Ryder C, Byrd M, Wozniak DJ (2007) Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol 10:644–648
Sabra W, Kim E-J, Zeng A-P (2002) Physiological responses of Pseudomonas aeruginosa PAO1 to oxidative stress in controlled microaerobic and aerobic cultures. Microbiol 148:3195–3202
Spangenberg C, Fislage R, Sierralta W, Tiimmler B, Ramling Ute (1995) Comparison of type IV-pilin genes of Pseudomonas aeruginosa of various habitats has uncovered a novel unusual sequence. FEMS Microbiol Lett 125:265–274
Stepanovic S, Vukovic D, Dakic I, Savic B, Svabic-Vlahovic M (2000) A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods 40(2):175–179
Sutherland IW (2001) The biofilm matrix—an immobilized but dynamic microbial environment. Trends Microbiol 9:222–227
Weger LA, Van der Vlught CIM, Wijfjes AHM, Bakker PAHM, Schippers B, Lugtenberg B (1987) Flagella of a plant-growth-stimulating Pseudomonas fluorescens are required for colonization of potato roots. J Bacteriol 169:2769–2773
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS (2002) Extracellular DNA required for bacterial biofilm formation. Science 295:1487
Acknowledgements
The authors are grateful to Mr Iiyama Kazuhiro of the Fukuoka University of Agriculture for providing bacterial strains.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Jorge Membrillo-Hernández.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Raouia, H., Hamida, B., Khadidja, A. et al. Effect of static magnetic field (200 mT) on biofilm formation in Pseudomonas aeruginosa. Arch Microbiol 202, 77–83 (2020). https://doi.org/10.1007/s00203-019-01719-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-019-01719-8