Abstract
Electronic Health Records (EHR) are inherently complex and diverse and cannot be readily integrated and analyzed. Analogous to the Gene Ontology, controlled clinical terminologies were created to facilitate the standardization and integration of medical concepts and knowledge and enable their subsequent use for translational research, official statistics and medical billing. This chapter will introduce several of the main controlled clinical terminologies used to record diagnoses, surgical procedures, laboratory results and medications. The discovery of novel therapeutic agents and treatments for rare or common diseases increasingly requires the integration of genotypic and phenotypic knowledge across different biomedical data sources. Mechanisms that facilitate this linkage, such as the Human Phenotype Ontology, are also discussed.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
1 Introduction
We are arguably entering the era of data-driven, personalized medicine, where electronic health records are considered the transformational force for measuring and improving the quality of clinical care and accelerating the pace of biomedical research [1, 2]. Electronic Health Record (EHR) data, alternatively referred to as Electronic Medical Record (EMR) data, are broadly defined as electronic data that are generated, captured and collected as part of routine clinical care across primary, secondary, and tertiary health care settings. EHR data can be structured (i.e., recorded using clinical terminologies), semi-structured (e.g., laboratory test results), or unstructured (e.g., free text). EHR data present multiple opportunities that have the potential to transform medical practice and research across all stages of translation [3–6].
Health care is an intrinsically multidisciplinary process and the care of patients, even within a single clinical specialty, intimately involves clinicians from a diverse set of other specialties (e.g., physicians, surgeons, radiologists, pharmacologists). Patient interactions often occur within distinct health care settings: some diseases are almost exclusively managed in primary care while acute manifestations are usually treated in secondary care. For chronic conditions, such as cardiovascular diseases, patients may have multiple interactions within primary and secondary care, and undergo assessments and diagnostic tests across both settings over long periods of time. The amount of EHR data being digitally generated and collected are thus vast and rapidly expanding but lack a common structure to facilitate their use, both for care across clinical settings but also for research, auditing, and other administrative purposes.
The purpose of this chapter is to provide a brief introduction to clinical terminologies for capturing and representing different aspects of clinical care in electronic health records. Firstly, contemporary terminologies for recording diagnoses, surgical procedures, lab measurements, and medication are described. Secondly, the main applications and challenges of using clinical terminologies are set out. Lastly, a potential pathway for integrating clinical terminologies with biological ontologies is illustrated through a case study in breast cancer.
2 Controlled Clinical Terminologies
Similar to bio-ontologies, such as the Gene Ontology [7, 8], controlled clinical terminologies (Table 1) were created to facilitate the systematic capture, curation, and description of health care-related concepts encountered during clinical care [9]. These can include but are not limited to diagnoses, symptoms, anatomical terms of location, prescribed medications, medical tests, surgical procedures, and laboratory measurements. Clinical terminologies are considered the conceptual core of clinical information systems and an essential tool for facilitating clinical data integration and reuse amongst disparate data sources. Initiatives such as the Open Biomedical Ontologies Consortium (OBO) [10] were founded to coordinate their evolution and alignment and provide a set of guidelines for creating and maintaining them with the aim of establishing an ecosystem of interoperable entities.
Several systematic literature reviews provide in-depth detail on their different aspects and characteristics [11–16]. A brief description of some key terminologies is provided below.
2.1 Diagnoses
SNOMED-Clinical Terms (SNOMED-CT) [17, 18] contains representations for over 300,000 health care-related concepts and is designed to capture and represent patient data for clinical care. It consists of four primary components that define the structure of the recorded information: concepts, descriptions, relationships and reference sets. Concepts are the basic unit of describing health care-related information and are uniquely identified, e.g., the Myocardial Infarction concept (id 22298006). All concepts have a unique Fully Specified Name, a list of Preferred Terms (e.g., Myocardial Infarction), and Synonyms (e.g., Heart attack, Cardiac infarction) defined. Concepts are organized into an acyclic hierarchy of is-a relationships that enables multiple inheritance i.e. concepts can have multiple parent concepts. For example Myocardial Infarction (id 22298006) is a subclass of the concepts Necrosis of anatomical site (id 609410002), Ischaemic heart disease (414545008), and Myocardial disease (id 57809008). SNOMED-CT contains terms for describing clinical findings, symptoms, diagnoses, procedures, medication, devices and anatomical body structures. It provides a compositional syntax which allows multiple ontology terms to be combined in order to build composite terms to represent complex medical concepts, a process known as post-coordination. Significant variation exists internationally with regards to SNOMED-CT adoption and implementation [19] and its use for research or routine clinical care. In the UK National Health Service (NHS), SNOMED-CT has been designated to become the standard clinical terminology to be used across the entire health care system by 2020.
The International Statistical Classification of Diseases and Related Health Problems (ICD) is a statistical classification system maintained by the World Health Organization [20]. ICD encapsulates concepts for classifying diseases, signs and symptoms, abnormal investigation findings, complaints, interactions with the health care system, social circumstances, and external causes of injury or disease. It maps health conditions to corresponding generic categories together with specific variations, assigning for these a designated alphanumeric code, up to six characters long. Major categories are designed to include a set of similar diseases (e.g., ICD chapter “I” encapsulates all diseases of the circulatory system). It is currently the most widely used statistical classification system in the world with many countries developing their own extensions and modifications tailored to their local health care system (e.g., ICD-9-CM used in the USA [21]). The primary use case of ICD is to abstract EHR data by assigning unique codes to diagnoses and procedures. This process is known as clinical coding, and performed manually or algorithmically by specialist staff according to a prespecified protocol. Coded data are then utilized for research [22], official statistics [23], medical billing, and health care resource planning.
2.2 Procedures
Clinical terminologies are used for describing surgical procedures, interventions, and investigations that patients undergo in hospitals, during in patient and outpatient interactions. In the USA, the American Medical Association maintains the Current Procedural Terminology [24] (CPT) and in the UK, the OPCS Classification of Interventions and Procedures version 4 (OPCS-4) [25] is used by the National Health Service. Both terminologies are used to convey information with regards to procedures to physicians and clinical coders and are combined with diagnosis codes during the medical billing process.
2.3 Laboratory Measurements
Logical Observation Identifiers Names and Codes (LOINC) [26–28] is maintained by the Regenstrief Institute and used for describing medical laboratory observations. LOINC facilitates the exchange of information with regards to laboratory tests and results between health care providers, laboratories and public health agencies. LOINC terms correspond to a single test, panel, observation, or measurement and are uniquely identified by a numeric code. Terms are formed of six parts: component (what is being measured), property (characteristics of what is being measured), time (measurement temporal information), system (observation context or specimen type), scale (scale of measure), and method (procedure used to obtain the measure).
2.4 Medication
RxNorm [29] is a US-specific terminology developed by the Library of Medicine for describing information about clinical drugs (defined as pharmaceutical products taken by patients with a therapeutic or diagnostic intent). It provides normalized names for all clinical drugs and links information about their active ingredient(s), strengths, form, and branded versions. RxNorm is widely used for recording drug information in patient health records, exchanging information between health care providers [30], personal medication records [31], and medication-related clinical decision support [32] and contains cross-references to other commonly used drug vocabularies.
3 Uses of Clinical Terminologies
While clinical terminologies are primarily used for the purposes of clinical data standardization and integration, the provision of a systematic and common language for describing health care concepts enables the subsequent use of EHR data for a diverse set of purposes, such as clinical research, auditing and billing. Adoption of clinical terminologies worldwide varies across health care settings and by purpose but diagnostic and procedural classification systems are primarily used for medical billing purposes. This section will briefly describe the opportunities and challenges of using EHR data and clinical terminologies.
3.1 Opportunities
EHR data are increasingly being linked and used for translational research [33] as they offer larger sample sizes at a higher clinical resolution [34]. A primary use-case of linked EHR data is to accurately extract phenotypic information (i.e., disease status), a process known as phenotyping [35]. Identifying cohorts of patients that share a common characteristic (e.g., have been diagnosed with hypertension or have abnormally high blood glucose measurements) enables researchers to use EHR data to perform large-scale clinical research studies at a lower cost compared to traditional bespoke investigator-led studies. EHR data have been used to examine disease aetiology in relation to clinical risk factors [36, 37] or genotypic information [38, 39], develop disease prognosis models [40], perform health outcome comparisons between countries [41], and facilitate pragmatic clinical trials [24]. Clinical terminologies are heavily used by deterministic rule-based algorithms curated by experts for identifying and constructing patient cohorts from raw EHR data but data-driven methodologies are increasingly being utilized [42]. Comprehensive reviews provide additional information on the use of clinical terminologies for other purposes such as annotating and accessing medical knowledge sources, data integration, semantic interoperability, data aggregation, and clinical decision support systems [43–46].
3.2 Challenges
Merging EHR data across sources becomes challenging due to the differences in the manner in which data are recorded. Each health care setting generates and records data for a particular purpose using the clinical terminology that is optimal in that specific context. For example, information in primary care can be recorded using SNOMED-CT whereas hospital morbidities would be recorded using ICD-10. This mismatch between the clinical terminologies used to record information leads to significant challenges as information is recorded at varying levels of granularity across sources. Semantic mapping systems, such as the Unified Medical Language System [47] (UMLS), can provide further details on the relationship between terms in each clinical terminology and facilitate the translation or integration of information across sources. However, direct one-to-one mappings might not always exist between terminologies leading to information loss due to insufficient resolution or conflicts between two sources where multiple potential mappings exist. These issues and their severity vary by clinical speciality and context but often require a set of rules to be created by users and manually applied in order to resolve them before the data can be used for research purposes. In cases of incomplete mappings, synonyms or adjacent terms in the clinical terminology might be used as a replacement term but that is assessed on a case-by-case basis.
4 Integrating Biological and Clinical Data
A key challenge in genomics is to understand and elucidate the phenotypic consequences of variation observed in the genotypic level. Even among Mendelian diseases, the association between genotype and phenotype is often complex. With the advent of next-generation sequencing methods, the focus is now shifting from generating genomic sequence data to efficiently interpreting them.
From a clinical care perspective, diseases presented by patients can be phenotypically distinct and associated with a specific set of treatments, symptoms, investigative procedures and management strategies. From a molecular scientist’s perspective however, it might be appropriate to group and analyze diseases that share a common biological pathway as a single entity in order to discover similarities in the way they manifest in different patient groups. Both of these viewpoints are valid, but as a direct consequence, data describing phenotypic and molecular properties are recorded in a different, and often incompatible, manner [48]. The problem is exacerbated in rare diseases where researchers are required to create larger cohorts of patients by pooling data across research consortia in order to increase the sample sizes and obtain accurate estimates of risk.
Increasing amounts of molecular function knowledge are being recorded in a hierarchical manner, using bio-ontologies such as the GO, which offer a rigid way to represent knowledge in a machine-readable manner, interoperable between different data sources and annotated [11]. Scientists aim to link and integrate this with phenotypic information in order to elucidate the genotype-phenotype relationship and facilitate the discovery of novel therapeutic agents and treatments for common or rare disorders. Ontologies such as the Human Phenotype Ontology (HPO) [49, 50] and the Disease Ontology [51, 52] were created to provide streamlined disease definitions by systematically combining the diverse and heterogeneous knowledge contained within clinical terminologies and other annotation sources under a single framework. These tools aim to provide researchers with a rich resource that semantically links diverse disease definitions from clinical terminologies and enables the linking of phenotypic, genotypic and genetic information of a disease.
4.1 Human Phenotype Ontology
The HPO is a structured, curated ontology describing phenotypic abnormalities and the relationships between them. The HPO aims to act as scaffolding for enabling the interoperability between molecular biology and human disease by providing a centralized resource for integrating genotypic and phenotypic data across biomedical sources. The HPO enables the computational analysis of human (and model organism) phenotypes against the background biological and molecular knowledge incorporated in biological ontologies such as the GO.
The HPO is organized as three independent sub-ontologies that cover different domains with the largest one being the one describing phenotypic abnormalities. The other two sub-ontologies describe the mode of inheritance and the onset and clinical course of the abnormalities. The primary focus of the HPO is not to capture diseases but rather the phenotypic abnormalities that are associated with them. Each HPO term describes a phenotypic abnormality (e.g., Primary congenital glaucoma) and is assigned a unique persistent identifier (e.g., HP:0001087). HPO terms are related to parent terms by “is a” relationships and terms can have multiple parent terms. The HPO is not primarily designed to capture and document quantitative information (e.g., systolic blood pressure, body mass index) but does provide qualitative descriptions of excess or reduction in quantity leading to a phenotypic abnormality (e.g., markedly reduced T cell function).
Interoperability between molecular and phenotypic data and research areas is accomplished through a comprehensive set of term annotations. The majority of HPO terms contain a reference to the Unified Medical Language System [47], enabling the mapping of terms between controlled clinical terminologies and other sources in the UMLS Metathesaurus. Additionally, HPO terms contain annotations that provide pointers to specific diseases or genes created in other external knowledge sources such as Online Mendelian Inheritance in Man (OMIM) database (http://omim.org/), DECIPHER (https://decipher.sanger.ac.uk/), and Orphanet (http://www.orpha.net/). HPO annotations have a number of metadata fields associated with them for further specifying onset, frequency and quantifying modifier effects. Annotations evidence codes, analogous to GO Evidence Codes, describe the manner in which a particular annotation was assigned to a term (e.g., inferred by text mining, traceable author statement, inferred from electronic annotation, public clinical study).
4.2 From Base Pairs to Bedside Phenotypes: Breast Cancer Case Study
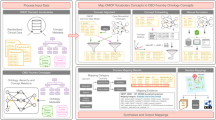
Using malignant neoplasms of the breast as a hypothetical case study, this section presents a potential pathway of linking biological knowledge on genotypic variation and molecular functions to clinical phenotypes encountered within the health care system. Drilling down from the right-hand side of clinical phenotypes down to the left-hand side of genotypic variation,
Figure 1 illustrates details of all potential sources and annotation mechanisms used within each source to capture and record information.
Along one potential path from genomic variation to genotypic information, transcripts and phenotypic information observed in clinical care there are multiple annotation mechanisms that are being utilized to record information in a structured way and enable the machine-driven interoperability between different platforms
Genotypic information: HPO annotations provide a cross-link to the Online Mendelian Inheritance in Man (OMIM) Breast Cancer, Familial phenotype entity (OMIM #114480—URL www.omim.org/entry/114480). OMIM provides curated lists of disease phenotypes and genes associated with that phenotype, in this case for example the BRCA2 gene entry (OMIM *600185—www.omim.org/entry/600185). Additionally, entries provide cross-links with Entrez [53] (Gene ID 675—URL http://www.ncbi.nlm.nih.gov/gene/675) and Ensembl [54] (ENSG00000139618—URL http://www.ensembl.org/Homo_sapiens/Gene/Summary?g=ENSG00000139618;r=13:32315474-32400266). Breast Cancer 2, early onset (BRCA2) is a protein-coding gene and belongs to the Fanconi anemia, complementation group (FANC) family of genes.
Genotypic variation: The NCBI dbSNP (http://www.ncbi.nlm.nih.gov/SNP/) provides curated and annotated information linking Single Nucleotide Polymorphisms (SNPs) and individual genes. rs144848 is one of the multiple mutations in the BRCA2 gene that have been reported to represent an independently minor but cumulatively significant increased risk for developing breast cancer [55]. dbSNP provides information the SNPs location (e.g., chromosome and chromosomal position), source assays, discordant genotypes and population diversity. (URL http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=rs144848)
Molecular function: UniProt [56] provides information on gene transcripts, in this case BRCA2_HUMAN (P51587, Breast cancer type 2 susceptibility protein). The biological process and molecular functions of the gene product are annotated using the Gene Ontology: double-strand break repair via homologous recombination (GO:0000724), DNA Repair (GO:0006281), cytokinesis (GO:0000910), protease binding (GO:0002020), and positive regulation of transcription, DNA-templated (GO:0045893). Using the GO, researchers are able to identify other gene products that share a common biological pathway or molecular function and incorporate that knowledge in their experiments. (URL: http://www.uniprot.org/uniprot/P51587)
Phenotypic information: The HPO Breast carcinoma term (HP: 0003002—http://purl.obolibrary.org/obo/HP_0003002) defines the presence of a carcinoma of the breast and is a child node of Neoplasms of the breast (HP:0100013). The HPO term contains a cross-reference to the Unified Medical Language System (UMLS) Malignant Neoplasm of Breast (UMLS:C0006142—URL https://uts.nlm.nih.gov//metathesaurus.html#C0006142;0;1;CUI;2015AA;EXACT_MATCH;*;) Concept which in turns provides mappings to other major controlled clinical terminologies such as the International Classification of Diseases 10th revision (C50, Malignant neoplasm of breast—http://apps.who.int/classifications/icd10/browse/2010/en#/C50-C50) and SNOMED-Clinical Terms (254837009, Malignant tumor of breast—http://bioportal.bioontology.org/ontologies/SNOMEDCT?p=classes&conceptid=254837009).
Clinical phenotype: Oncology data in hospitals are stored in diverse locations and formats since diagnosis and treatment is a multidisciplinary process between pathology, radiology, surgery, medical oncology and radiotherapy. Breast cancer diagnosis and severity is usually evaluated through imaging tests such as mammograms, ultrasounds, magnetic resonance imaging or by performing a biopsy. Medical images and their associated metadata are stored in a picture archiving and communication system (PACS) system and information about these procedures and the results obtained would be recorded using intervention and procedure terms. Diagnosis and staging information would be stored and coded in pathology systems using a medical terminology such as SNOMED-CT or other bespoke data structures. Treatment data would be stored in the pharmacy information systems.
5 Conclusion
The amount of clinical data that are generated and captured during routine clinical care is increasing in size and complexity. Integrating clinical data from disparate sources however is a challenging task due to their lack of common structure and annotation. Similar to the Gene Ontology, controlled clinical terminologies have been created to facilitate the systematic capture, curation, and description of health care related events such as diagnoses, prescriptions and procedures from EHR data and enable their subsequent usage for clinical care, research, or administrative purposes. Furthermore, linking EHR data with biological knowledge is increasingly becoming possibly through tools such as the Human Phenotype Ontology (HPO) and the Disease Ontology that aim to provide the semantic scaffolding for computationally integrating biomedical knowledge across sources.
Funding Open Access charges were funded by the University College London Library, the Swiss Institute of Bioinformatics, the Agassiz Foundation, and the Foundation for the University of Lausanne.
References
Collins F, Varmus H (2015) A New Initiative on Precision Medicine. N Engl J Med 372(9):793–795. doi:10.1056/nejmp1500523
Shah N, Tenenbaum J (2012) The coming age of data-driven medicine: translational bioinformatics’ next frontier. J Am Med Inform Assoc 19:e1. doi:10.1136/amiajnl-2012-000969
Jensen P, Jensen L, Brunak S (2012) Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet 13(6):395–405. doi:10.1038/nrg3208
Khoury M, Gwinn M, Ioannidis J (2010) The emergence of translational epidemiology: from scientific discovery to population health impact. Am J Epidemiol 172(5):517–524. doi:10.1093/aje/kwq211
Khoury M, Lam TK, Ioannidis J, Hartge P, Spitz M, Buring J, Chanock S, Croyle R, Goddard K, Ginsburg G, Herceg Z, Hiatt R, Hoover R, Hunter D, Kramer B, Lauer M, Meyerhardt J, Olopade O, Palmer J, Sellers T, Seminara D, Ransohoff D, Rebbeck T, Tourassi G, Winn D, Zauber A, Schully S (2013) Transforming epidemiology for 21st century medicine and public health. Cancer Epidemiol Biomark Prev 22(4):508–516. doi:10.1158/1055-9965.epi-13-0146
Liao K, Cai T, Savova G, Murphy S, Karlson E, Ananthakrishnan A, Gainer V, Shaw S, Xia Z, Szolovits P, Churchill S, Kohane I (2015) Development of phenotype algorithms using electronic medical records and incorporating natural language processing. BMJ 350:h1885. doi:10.1136/bmj.h1885
Ashburner M, Ball C, Blake J, Botstein D, Butler H, Cherry M, Davis A, Dolinski K, Dwight S, Eppig J, Harris M, Hill D, Issel-Tarver L, Kasarskis A, Lewis S, Matese J, Richardson J, Ringwald M, Rubin G, Sherlock G (2000) Gene Ontology: tool for the unification of biology. Nat Genet 25(1):25–29. doi:10.1038/75556
Gene Ontology C (2004) The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res 32(Suppl 1):D258–D261. doi:10.1093/nar/gkh036
Cantor M, Lussier Y (2003) Putting data integration into practice: using biomedical terminologies to add structure to existing data sources. AMIA Annual Symposium proceedings/AMIA Symposium AMIA Symposium, pp 125–129
Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg L, Eilbeck K, Ireland A, Mungall C, Leontis N, Rocca-Serra P, Ruttenberg A, Sansone S-A, Scheuermann R, Shah N, Whetzel P, Lewis S (2007) The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol 25(11):1251–1255. doi:10.1038/nbt1346
Bard J, Rhee S (2004) Ontologies in biology: design, applications and future challenges. Nat Rev Genet 5(3):213–222. doi:10.1038/nrg1295
Cimino JJ (1998) Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med 37(4-5):394–403
Chute CG, Cohn SP, Campbell KE, Oliver DE, Campbell JR (1996) The content coverage of clinical classifications. For The Computer-Based Patient Record Institute’s Work Group on Codes & Structures. J Am Med Inform Assoc 3(3):224–233
Pathak J, Wang J, Kashyap S, Basford M, Li R, Masys D, Chute C (2011) Mapping clinical phenotype data elements to standardized metadata repositories and controlled terminologies: the eMERGE Network experience. J Am Med Inform Assoc 18(4):376–386. doi:10.1136/amiajnl-2010-000061
de Lusignan S, Minmagh C, Kennedy J, Zeimet M, Bommezijn H, Bryant J (2001) A survey to identify the clinical coding and classification systems currently in use across Europe. Stud Health Technol Inform 84(Pt 1):86–89
de Lusignan S (2005) Codes, classifications, terminologies and nomenclatures: definition, development and application in practice. Inform Prim Care 13(1):65–70
Donnelly K (2006) SNOMED-CT: the advanced terminology and coding system for eHealth. Stud Health Technol Inform 121:279–290
Wang A, Sable J, Spackman K (2002) The SNOMED clinical terms development process: refinement and analysis of content. Proceedings/AMIA Annual Symposium AMIA Symposium, pp 845–849
Lee D, de Keizer N, Lau F, Cornet R (2014) Literature review of SNOMED CT use. J Am Med Inform Assoc 21:e1. doi:10.1136/amiajnl-2013-001636
Denny J, Crawford D, Ritchie M, Bielinski S, Basford M, Bradford Y, Chai HS, Bastarache L, Zuvich R, Peissig P, Carrell D, Ramirez A, Pathak J, Wilke R, Rasmussen L, Wang X, Pacheco J, Kho A, Hayes G, Weston N, Matsumoto M, Kopp P, Newton K, Jarvik G, Li R, Manolio T, Kullo I, Chute C, Chisholm R, Larson E, McCarty C, Masys D, Roden D, de Andrade M (2011) Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet 89(4):529–542. doi:10.1016/j.ajhg.2011.09.008
Quan H, Sundararajan V, Halfon P, Fong A, Burnand B, Luthi J-C, Saunders D, Beck C, Feasby T, Ghali W (2005) Coding algorithms for defining comorbidities in ICD-9-CM and ICD-10 administrative data. Med Care 43(11):1130–1139
Rubbo B, Fitzpatrick N, Denaxas S, Daskalopoulou M, Yu N, Patel R, Hemingway H (2015) Use of electronic health records to ascertain, validate and phenotype acute myocardial infarction: a systematic review and recommendations. Int J Cardiol. doi:10.1016/j.ijcard.2015.03.075
Taylor P (2013) Standardized mortality ratios. Int J Epidemiol 42(6):1882–1890. doi:10.1093/ije/dyt209
van Staa T-P, Dyson L, McCann G, Padmanabhan S, Belatri R, Goldacre B, Cassell J, Pirmohamed M, Torgerson D, Ronaldson S, Adamson J, Taweel A, Delaney B, Mahmood S, Baracaia S, Round T, Fox R, Hunter T, Gulliford M, Smeeth L (2014) The opportunities and challenges of pragmatic point-of-care randomised trials using routinely collected electronic records: evaluations of two exemplar trials. Health Technol Assess (Winchester, England) 18(43):1–146
Scannell J, Blanckley A, Boldon H, Warrington B (2012) Diagnosing the decline in pharmaceutical R&D efficiency. Nat Rev Drug Discov 11(3):191–200. doi:10.1038/nrd3681
Bakken S, Cimino JJ, Haskell R, Kukafka R, Matsumoto C, Chan GK, Huff SM (2000) Evaluation of the clinical LOINC (Logical Observation Identifiers, Names, and Codes) semantic structure as a terminology model for standardized assessment measures. J Am Med Inform Assoc 7(6):529–538
Huff SM, Rocha RA, McDonald CJ, De Moor GJ, Fiers T, Bidgood WD, Forrey AW, Francis WG, Tracy WR, Leavelle D, Stalling F, Griffin B, Maloney P, Leland D, Charles L, Hutchins K, Baenziger J (1998) Development of the Logical Observation Identifier Names and Codes (LOINC) vocabulary. J Am Med Inform Assoc 5(3):276–292
McDonald C, Huff S, Suico J, Hill G, Leavelle D, Aller R, Forrey A, Mercer K, DeMoor G, Hook J, Williams W, Case J, Maloney P (2003) LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clin Chem 49(4):624–633. doi:10.1373/49.4.624
Nelson S, Zeng K, Kilbourne J, Powell T, Moore R (2011) Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc 18(4):441–448. doi:10.1136/amiajnl-2011-000116
Bouhaddou O, Warnekar P, Parrish F, Do N, Mandel J, Kilbourne J, Lincoln M (2008) Exchange of computable patient data between the Department of Veterans Affairs (VA) and the Department of Defense (DoD): terminology mediation strategy. J Am Med Inform Assoc 15(2):174–183
Zeng K, Bodenreider O, Nelson S (2008) Design and implementation of a personal medication record-MyMedicationList. AMIA Annual Symposium proceedings/AMIA Symposium AMIA Symposium, pp 844–848
Duke J, Friedlin J (2010) ADESSA: a real-time decision support service for delivery of semantically coded adverse drug event data. AMIA Annu Symp Proc 2010:177–181
Weber G, Mandl K, Kohane I (2014) Finding the missing link for big biomedical data. JAMA. doi:10.1001/jama.2014.4228
Denaxas S, Morley K (2015) Big biomedical data and cardiovascular disease research: opportunities and challenges. Eur Heart J 1(1): qcv005. doi: 10.1093/ehjqcco/qcv005
Morley K, Wallace J, Denaxas S, Hunter R, Patel R, Perel P, Shah A, Timmis A, Schilling R, Hemingway H (2014) Defining disease phenotypes using national linked electronic health records: a case study of atrial fibrillation. PLOS ONE 9(11):e110900
Rapsomaniki E, Timmis A, George J, Pujades-Rodriguez M, Shah A, Denaxas S, White I, Caulfield M, Deanfield J, Smeeth L, Williams B, Hingorani A, Hemingway H (2014) Blood pressure and incidence of twelve cardiovascular diseases: lifetime risks, healthy life-years lost, and age-specific associations in 1·25 million people. Lancet 383(9932):1899–1911
Shah AD, Langenberg C, Rapsomaniki E, Denaxas S, Pujades-Rodriguez M, Gale C, Deanfield J, Smeeth L, Timmis A, Hemingway H (2015) Type 2 diabetes and incidence of cardiovascular diseases: a cohort study in 1·9 million people. Lancet Diabetes Endocrinol 3(2):105–113
Newton K, Peissig P, Kho A, Bielinski S, Berg R, Choudhary V, Basford M, Chute C, Kullo I, Li R, Pacheco J, Rasmussen L, Spangler L, Denny J (2013) Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc 20(e1):e147–e154. doi:10.1136/amiajnl-2012-000896
Gottesman O, Kuivaniemi H, Tromp G, Faucett A, Li R, Manolio T, Sanderson S, Kannry J, Zinberg R, Basford M, Brilliant M, Carey D, Chisholm R, Chute C, Connolly J, Crosslin D, Denny J, Gallego C, Haines J, Hakonarson H, Harley J, Jarvik G, Kohane I, Kullo I, Larson E, McCarty C, Ritchie M, Roden D, Smith M, Böttinger E, Williams M (2013) The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med 15(10):761–771. doi:10.1038/gim.2013.72
Rapsomaniki E, Shah A, Perel P, Denaxas S, George J, Nicholas O, Udumyan R, Feder G, Hingorani A, Timmis A, Smeeth L, Hemingway H (2013) Prognostic models for stable coronary artery disease based on electronic health record cohort of 102 023 patients. Eur Heart J:eht533. doi:10.1093/eurheartj/eht533
Chung S-C, Gedeborg R, Nicholas O, James S, Jeppsson A, Wolfe C, Heuschmann P, Wallentin L, Deanfield J, Timmis A, Jernberg T, Hemingway H (2014) Acute myocardial infarction: a comparison of short-term survival in national outcome registries in Sweden and the UK. Lancet 383(9925):1305–1312. doi:10.1016/s0140-6736(13)62070-x
Shivade C, Raghavan P, Fosler-Lussier E, Embi P, Elhadad N, Johnson S, Lai A (2014) A review of approaches to identifying patient phenotype cohorts using electronic health records. J Am Med Inform Assoc 21(2):221–230. doi:10.1136/amiajnl-2013-001935
Denny J (2012) Chapter 13: Mining electronic health records in the genomics era. PLoS Comput Biol 8(12):e1002823. doi:10.1371/journal.pcbi.1002823
Bodenreider O (2008) Biomedical ontologies in action: role in knowledge management, data integration and decision support. Yearb Med Inform 2008:67–79
Bodenreider O, Stevens R (2006) Bio-ontologies: current trends and future directions. Brief Bioinform 7(3):256–274. doi:10.1093/bib/bbl027
Stanfill M, Williams M, Fenton S, Jenders R, Hersh W (2010) A systematic literature review of automated clinical coding and classification systems. J Am Med Inform Assoc 17(6):646–651. doi:10.1136/jamia.2009.001024
Bodenreider O (2004) The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res 32(Suppl 1):D267–D270. doi:10.1093/nar/gkh061
Biesecker L (2004) Phenotype matters. Nat Genet 36(4):323–324. doi:10.1038/ng0404-323
Robinson P, Köhler S, Bauer S, Seelow D, Horn D, Mundlos S (2008) The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am J Hum Genet 83(5):610–615. doi:10.1016/j.ajhg.2008.09.017
Köhler S, Doelken S, Mungall C, Bauer S, Firth H, Bailleul-Forestier I, Black G, Brown D, Brudno M, Campbell J, FitzPatrick D, Eppig J, Jackson A, Freson K, Girdea M, Helbig I, Hurst J, Jähn J, Jackson L, Kelly A, Ledbetter D, Mansour S, Martin C, Moss C, Mumford A, Ouwehand W, Park S-M, Riggs E, Scott R, Sisodiya S, Van Vooren S, Wapner R, Wilkie A, Wright C, Vulto-van Silfhout A, de Leeuw N, de Vries B, Washingthon N, Smith C, Westerfield M, Schofield P, Ruef B, Gkoutos G, Haendel M, Smedley D, Lewis S, Robinson P (2014) The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res 42(D1):D966–D974. doi:10.1093/nar/gkt1026
Osborne J, Flatow J, Holko M, Lin S, Kibbe W, Zhu L, Danila M, Feng G, Chisholm R (2009) Annotating the human genome with Disease Ontology. BMC Genomics 10(Suppl 1):S6. doi:10.1186/1471-2164-10-s1-s6
Schriml LM, Arze C, Nadendla S, Chang Y-WW, Mazaitis M, Felix V, Feng G, Kibbe WA (2012) Disease Ontology: a backbone for disease semantic integration. Nucleic Acids Res 40(Database issue):D940–D946. doi:10.1093/nar/gkr972
Maglott D, Ostell J, Pruitt K, Tatusova T (2005) Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res 33(Suppl 1):D54–D58. doi:10.1093/nar/gki031,
Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, Durbin R, Eyras E, Gilbert J, Hammond M, Huminiecki L, Kasprzyk A, Lehvaslaiho H, Lijnzaad P, Melsopp C, Mongin E, Pettett R, Pocock M, Potter S, Rust A, Schmidt E, Searle S, Slater G, Smith J, Spooner W, Stabenau A, Stalker J, Stupka E, Ureta-Vidal A, Vastrik I, Clamp M (2002) The Ensembl genome database project. Nucleic Acids Res 30(1):38–41. doi:10.1093/nar/30.1.38
Johnson N, Fletcher O, Palles C, Rudd M, Webb E, Sellick G, dos Santos SI, McCormack V, Gibson L, Fraser A, Leonard A, Gilham C, Tavtigian S, Ashworth A, Houlston R, Peto J (2007) Counting potentially functional variants in BRCA1, BRCA2 and ATM predicts breast cancer susceptibility. Hum Mol Genet 16(9):1051–1057. doi:10.1093/hmg/ddm050
Apweiler R, Bairoch A, Wu C, Barker W, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin M, Natale D, O’Donovan C, Redaschi N, Yeh LS (2004) UniProt: the Universal Protein knowledgebase. Nucleic Acids Res 32(Suppl 1):D115–D119. doi:10.1093/nar/gkh131
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
This chapter is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, duplication, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, a link is provided to the Creative Commons license and any changes made are indicated.
The images or other third party material in this chapter are included in the work’s Creative Commons license, unless indicated otherwise in the credit line; if such material is not included in the work’s Creative Commons license and the respective action is not permitted by statutory regulation, users will need to obtain permission from the license holder to duplicate, adapt or reproduce the material.
Copyright information
© 2017 The Author(s)
About this protocol
Cite this protocol
Denaxas, S.C. (2017). Integrating Bio-ontologies and Controlled Clinical Terminologies: From Base Pairs to Bedside Phenotypes. In: Dessimoz, C., Škunca, N. (eds) The Gene Ontology Handbook. Methods in Molecular Biology, vol 1446. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-3743-1_20
Download citation
DOI: https://doi.org/10.1007/978-1-4939-3743-1_20
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-3741-7
Online ISBN: 978-1-4939-3743-1
eBook Packages: Springer Protocols