Abstract
The Gene Ontology (GO) provides a framework and set of concepts for describing the functions of gene products from all organisms. It is specifically designed for supporting the computational representation of biological systems. A GO annotation is an association between a specific gene product and a GO concept, together making a statement pertinent to the function of that gene. However, the meaning of the term “function” is not as straightforward as it might seem, and has been discussed at length in both philosophical and biological circles. Here, I first review these discussions. I then present an explicit formulation of the biological model that underlies the GO and annotations, and discuss how this model relates to the broader debates on the meaning of biological function.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
1 What Is Biological Function?
The notion of function in biology has received a great deal of attention in the philosophical literature. At the broadest level, there are two schools of thought on how functions should be defined, now most commonly referred to as “causal role function” and “selected effect function.” Causal role function was first proposed by Cummins [1], and it focuses on describing function in terms of how a part contributes to some overall capacity of the system that contains the part. In this formulation, the function of an entity is relative to some system to which it contributes. For example, the statement “the function of the heart is to pump blood” has meaning only in the context of the larger circulatory system’s capacity to deliver nutrients and remove waste products from bodily tissues. However, one of the main objections to the causal role definition of function is that there is no systematic way to identify what the larger system (and the relevant capacity of that system) should be. Selected effect function, on the other hand, derives from the “etiological” definition of function first proposed by Wright [2]. In this formulation, a function of an entity is the ultimate answer to the question of why the entity exists at all. In biology, as explained by Millikan [3] and Neander [4], this is tantamount to asking the following: For which of its effects was it selected during evolution? One obvious advantage of the selected effect definition is that it explicitly incorporates evolutionary considerations, and demands that a function ultimately derive from its history of natural selection. On the more practical side, it has the further advantage of putting constraints on which effects, out of the myriad causal effects that a particular entity might have, could be considered as functions. Following the example above, an effect of the heart (beating) is to produce a sound, but it would not be correct to say that the function of the heart is to produce a sound. The selected effects definition of function would distinguish a proper function (e.g., pumping blood) from an “accidental” effect (e.g., producing a sound) on the basis that natural selection more likely operated on the heart’s effect of pumping blood. In the causal role definition, on the other hand, there is always the potential for arbitrariness and idiosyncrasy in defining a containing system and capacities; thus there is no general rule for distinguishing functional from accidental effects.
Nevertheless, causal role function has been stalwartly defended by biologists in the subdiscipline of functional anatomy [5], which emphasizes how anatomical parts function as parts of larger systems. They claim that the selected trait can be difficult to infer, and lack of a hypothesis for such a trait should not stand in the way of an analysis of the mechanism of how an anatomical feature operates. For example, one could analyze a jaw in terms of its capacity for generating a crushing force irrespective of whether it was selected for crushing seeds or defending against a predator. Indeed, the search for mechanisms of operation, or more generally just “mechanism,” has more recently been offered as an alternative paradigm for molecular and neurobiology in particular [6]. Mechanism, like causal role, focuses on how parts contribute to a system. But it takes a step further in defining core concepts, and how these relate to function. The core concepts are entities and activities: physical entities (such as proteins) perform activities, or actions that can have causal effects on other activities. In this view, a function is simply an activity that is carried out as part of a larger mechanism. For example, the function of the ribosome (an entity) is translation (an activity), and translation plays a role in a larger mechanism of gene expression. The subtle difference from earlier formulations of function is an emphasis on the activity having the role of a function, rather than the entity itself having a function. Also like causal role, no a priori constraints are put on mechanism: “a function is … a component in some mechanism, that is … in a context that is taken to be important, vital, or otherwise significant.” Clearly mechanism is susceptible to the same criticism as causal role function, regarding arbitrariness in the choice of system.
The core differences between selected effect function and causal role function derive largely from differences in what question they are trying to answer. For selected effect function, the question is about origins: Why is the entity there (i.e., what explains its selective advantage)? [2]. For causal role function, the question is about operation: How does the entity contribute to the biological capacities of the organism that has the entity (and only secondarily, how do those capacities relate to natural selection)? [1]. And there is little doubt that in most biological research endeavors today, the concern is in elucidating the mechanisms by which biological systems operate, rather than in explaining why the parts are there to begin with.
The notion of function, particularly in connection with molecular biology, has been discussed at length not only by philosophers, but also by molecular biologists themselves. As a representative sample, I will consider two publications written with very different aims in mind: a textbook chapter by Alberts entitled “Protein Function” [7] and a philosophical treatise by Monod, Chance and Necessity [8]. Alberts’ treatment of “function” covers two distinct but related senses of the word. The first is how an individual protein works at the mechanistic level (its manner of functioning): “how proteins bind to other selected molecules and how their activity depends on such binding.” The second is to describe how a protein acts as a component in a larger system, by analogy to mechanical parts in human-designed systems (its functional role in the context of the operation of the cell): “proteins … act as catalysts, signal receptors, switches, motors, or tiny pumps.” Specific molecular binding can be considered the general mechanism by which a functional role can be carried out. These uses of “function” appear, at least on the face of it, to be more in line with the causal role and mechanism views in the philosophical literature.
Given its broader intended audience of scientists and laymen (and presumably philosophers), Chance and Necessity puts biological function in a much broader context. Monod coins the term “teleonomic function” to describe more precisely what he means by function. He carefully defines teleonomy as the characteristic of “objects endowed with a purpose or project, which at the same time they exhibit through their structure and carry out through their performances” [p. 9]. Teleonomy is also a property of human-designed “artifacts,” further emphasizing the view of function in terms of an apparent purpose in accomplishing a predetermined aim. But living systems owe their teleonomy to a distinct source. As he so eloquently (if also compactly) states, “invariance necessarily precedes teleonomy” [p. 23], which he goes on to explain further as “the Darwinian idea that the initial appearance, evolution and steady refinement of ever more intensely teleonomic structures are due to perturbations in a structure which already possesses the property of invariance.” Thus what appears to be a future-goal-oriented action by a living organism is, in fact, only a blind repetition of a genetic program that evolved in the past. Importantly, Monod notes the presence of teleonomy at all levels of a biological system, from proteins (which he calls “the essential molecular agents of teleonomic performance”) to “systems providing large scale coordination of the organism’s performances … [such as] the endocrine and nervous systems” [p. 62]. In this way, Monod’s teleonomic function includes aspects of both Wright’s selected effect function (the origin of apparently designed functions in prior natural selection) and Cummins’s causal role function (the role of a part in a larger system).
In summary, function as conceived by molecular biologists (in what could be called the “molecular biology paradigm”) refers to specific, coordinated activities that have the appearance of having been designed for a purpose. That apparent purpose is their function. The appearance of design derives from natural selection, so many biologists now favor the use of the term “biological program” to avoid connotations of intentional design. Following this convention, biological programs, when executed, perform a function; that is, they result in a particular, previously selected outcome or causal effect. Biological programs are nested modularly inside other, larger biological programs, so a protein can be said to have functions at multiple levels. The lowest level biological program is expression of a single macromolecule, e.g., a protein: the gene is transcribed into RNA, which is translated into a protein, which adopts a particular structure that performs its function simply by following physical laws that determine how it will interact with specific (i.e., a small number) of other distinct types of other molecular entities. At higher levels, the functions of multiple proteins are executed in a coherent, controlled (“regulated”) manner to accomplish a larger function. Thus, simply identifying a coherent, regulated system of activities can be a fruitful, practical start for identifying selected effect functions. Causal role analyses can and do play such a role in functional anatomy and molecular biology. But of course they are only candidates for evolved biological functions until they have been related to past survival and reproduction, the ultimate function of every biological program.
2 Function in the Gene Ontology
I now turn to a description of how function is conceived of, and represented in practice, in the Gene Ontology.
2.1 Gene Products, Not Genes, Have Functions
In order to understand how gene function is represented in the GO, some basic molecular biology knowledge is required.
-
A gene is a contiguous region of DNA that encodes instructions for how the cell can make a large (“macro”) molecule (or potentially multiple different macromolecules).
-
A macromolecule is called a gene product (as it is produced deterministically according to the instructions from a gene), and can be of two types, a protein (the most common type) or a noncoding RNA.
-
A gene product can act as a molecular machine; that is, it can perform a chemical action that we call an activity.
-
Gene products from different genes can combine into a larger molecular machine, called a macromolecular complex.
Each concept in the Gene Ontology relates to the activity of a gene product or complex, as these are the entities that carry out cellular processes. A gene encodes a gene product, so it can obviously be considered the ultimate source of these activities and processes. But strictly speaking, a gene does not perform an activity itself. Thus, when the Gene Ontology refers to “gene function,” it is actually shorthand for “gene product function.”
2.2 Assertions About Functions of Particular Genes Are Made by “GO Annotations”
The Gene Ontology defines the “universe” of possible functions a gene might have, but it makes no claims about the function of any particular gene. Those claims are, instead, captured as “GO annotations.” A GO annotation is a statement about the function of a particular gene. But our biological knowledge is extremely incomplete. Accordingly, the GO annotation format is designed to capture partial, incomplete statements about gene function. A GO annotation typically associates only a single GO concept with a single gene. Together, these statements comprise a “snapshot” of current biological knowledge. Different pieces of knowledge regarding gene function may be established to different degrees, which is why each GO annotation always refers to the evidence upon which it is based.
2.3 The Model of Gene Function Underlying the GO
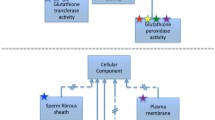
The Gene Ontology (GO) considers three distinct aspects of how gene functions can be described: molecular function, cellular component, and biological process (note that throughout this chapter, bold text will denote specific concepts, or classes, from the Gene Ontology). In order to understand what these aspects mean and how they relate to each other, it may be helpful to consider the biological model assumed in GO annotations. GO follows what could be called the “molecular biology paradigm,” as described in the previous section. In this representation, a gene encodes a gene product, and that gene product carries out a molecular-level process or activity (molecular function) in a specific location relative to the cell (cellular component), and this molecular process contributes to a larger biological objective (biological process) comprised of multiple molecular-level processes. An example, elaborating on the example in the original GO paper [9], is shown in Fig. 1.
To reiterate, GO concepts were designed to apply specifically to the actions of gene products, i.e., macromolecular machines comprising proteins, RNAs, and stable complexes thereof. In the GO representation, a region of DNA (e.g., a regulatory region) is treated not as carrying out a molecular process, but rather as an object that gene products can act upon in order to perform their specific activities.
2.4 Molecular Functions Define Molecular Processes (Activities)
In the GO, a molecular function is a process that can be carried out by the action of a single macromolecular machine, via direct physical interactions with other molecular entities. Function in this sense denotes an action, or activity, that a gene product performs. These actions are described from the two distinct but related perspectives commonly employed by biologists: (1) biochemical activity, and (2) role as a component in a larger system/process. Biochemical activities include binding and catalytic activities, and are only functions in the broad sense, i.e., how something functions, the molecular mechanism of operation. Component role descriptions, on the other hand, refer to roles in larger processes, and are sometimes described by analogy to a mechanical or electrical system. For example, biologists may refer to a protein that functions (acts) as a receptor. This is because the activity is interpreted as receiving a signal, and converting that signal into another physicochemical form. Unlike biochemical activities, these roles require some degree of interpretation that includes knowledge of the larger system context in which the gene product acts.
2.5 Cellular Components Define Places Where Molecular Processes Occur
A cellular component is a location, relative to cellular compartments and structures, occupied by a macromolecular machine when it carries out a molecular function. There are two ways in which biologists describe locations of gene products: (1) relative to cellular structures (e.g., cytoplasmic side of plasma membrane) or compartments (e.g., mitochondrion), and (2) the stable macromolecular complexes of which they are parts (e.g., the ribosome). Unlike the other aspects of GO, cellular component concepts refer not to processes but rather a cellular anatomy. Nevertheless, they are designed to be applied to the actions of gene products and complexes: a GO annotation to a cellular component provides information about where a molecular process may occur during a larger process.
2.6 Biological Processes Define Biological Programs Comprised of Regulated Molecular Processes
In the GO, a biological process represents a specific objective that the organism is genetically “programmed” to achieve. Each biological process is often described by its outcome or ending state, e.g., the biological process of cell division results in the creation of two daughter cells (a divided cell) from a single parent cell. A biological process is accomplished by a particular set of molecular processes carried out by specific gene products, often in a highly regulated manner and in a particular temporal sequence.
An annotation of a particular gene product to a GO biological process concept should therefore have a clear interpretation: the gene product carries out a molecular process that plays an integral role in that biological program. But a gene product can affect a biological objective even if it does not act strictly within the process, and in these cases a GO annotation aims to specify that relationship insofar as it is known. First, a gene product can control when and where the program is executed; that is, it might regulate the program. In this case, the gene product acts outside of the program, and controls (directly or indirectly) the activity of one or more gene products that act within the program. Second, the gene product might act in another, separate biological program that is required for the given program to occur. For instance, animal embryogenesis requires translation, though translation would not generally be considered to be part of the embryogenesis program. Thus, currently a given biological process annotation could have any of these three meanings (namely a gene activity could be part of, regulate, or be upstream of but still necessary for, a biological process). The GO Consortium is currently exploring ways to computationally represent these different meanings so they can be distinguished.
Biological process is the largest of the three ontology aspects in the GO, and also the most diverse. This reflects the multiplicity of levels of biological organization at which genetically encoded programs can be identified. Biological process concepts span the entire range of how biologists characterize biological systems. They can be as simple as a generic enzymatic process, e.g., protein phosphorylation, to molecular pathways such as glycolysis or the canonical Wnt signaling pathway, to complex programs like embryo development or learning, and even including reproduction, the ultimate function of every evolutionarily retained gene.
Because of this diversity, in practice not all biological process classes actually represent coherent, regulated biological programs. In particular, GO biological process also includes molecular-level processes that cannot always be distinguished from molecular functions. Taking the previous example, the process class protein phosphorylation overlaps in meaning with the molecular activity class protein kinase activity, as protein kinase activity is the enzymatic activity by which protein phosphorylation occurs. The main difference is that while a molecular function annotation has a precise semantics (e.g., the gene carries out protein kinase activity), the biological process annotation does not (e.g., the gene either carries out, regulates, or is upstream of but necessary for a particular protein kinase activity).
3 How Does the GO Relate to the Debate About the Meaning of Biological Function?
GO concepts are designed to describe aspects (molecular activity, location of the activity, and larger biological programs) of the functions that a gene evolved to perform, i.e., selected effect functions. However, GO concepts may not always be applied that way. As a result, a given GO annotation may or may not be a statement about selected effect function. Note that while all biological programs are carried out by molecular activities, not all molecular activities necessarily contribute to a biological program. In principle, then, only those GO annotations that refer to biological programs can be considered to generally reflect selected effect functions.
A GO molecular function annotation by itself cannot be automatically interpreted as selected effect function. One of the most vigorous long-standing debates in the GO Consortium concerns the protein binding class in GO, as it is clearly appreciated by biologists that a given experimental observation of molecular binding may reflect biological noise and not necessarily contribution to a biological objective. Even further removed, cellular component annotations are often made from observations of a protein in a particular compartment, irrespective of whether the protein performs a molecular activity in that location. For example, many proteins known to act extracellularly are also observed in the Golgi apparatus as they await trafficking to the plasma membrane. In short, if the molecular activity and cellular location are not yet implicated in a biological program (that is itself clearly related to survival and reproduction), they cannot be said to have selected effect function. Strictly speaking, such annotations should be considered as referring to candidate functions, rather than proper functions.
Despite these theoretical considerations, most GO annotations are likely in practice to refer to selected effect functions. This is simply because most GO annotations are made from publications describing specific, small-scale molecular biology studies that focus on a particular biological program. In such studies, a biological objective (usually implicitly related to survival and reproduction) has already been established in advance, and the paper describes the mechanistic activities of gene products in accomplishing that biological objective. Large-scale studies, on the other hand, that measure gene product activities or locations without reference to the biological program they are part of, should be considered as candidate selected effect functions. This view would address the recent debate about gene function [10–12], initiated when the ENCODE (Encyclopedia of DNA Elements) project—a large-scale, hypothesis-free project to catalog biochemical activities across numerous regions of the human genome [13]—inappropriately claimed to have discovered proper functions. The GO Consortium is discussing ways to help users distinguish between hypothesis-driven annotations (likely proper functions) from large-scale annotations (candidate functions).
4 Conclusion
It has not generally been appreciated that the Gene Ontology concepts for describing aspects of gene function assume a specific model of how gene products act to achieve biological objectives. My aim here has been to describe this model, which, I hope, will clarify how GO annotations should be properly used and interpreted, as well as how the GO relates to biological function as discussed in both the philosophical and biological literature.
References
Cummins R (1975) Functional analysis. J Philos 72:741–765
Wright L (1973) Functions. Philos Rev 82:139–168
Millikan RG (1989) In defense of proper functions. Philos Sci 56:288–302
Neander K (1991) The teleological notion of “function.”. Australas J Philos 69:454–468
Amundson R, Lauder GV (1994) Function without purpose: the uses of causal role function in evolutionary biology. In: Hull DL, Ruse M (eds) Biology & philosophy, vol 9. Oxford University Press, Oxford, pp 443–469
Machamer P, Darden L, Craver CF (2000) Thinking about mechanisms. Philos Sci 67:1–25
Alberts B (2002) Protein function. In: Molecular biology of the cell, 4th edn. Garland Science, New York
Monod J (1971) Chance and necessity. Alfred Knopf, New York
Ashburner MA et al (2000) Gene ontology: tool for the unification of biology. Nat Genet 25:25–29
Doolittle WF (2013) Is junk DNA bunk? A critique of ENCODE. Proc Natl Acad Sci U S A 110:5294–5300
Doolittle WF et al (2014) Distinguishing between “function” and “effect” in genome biology. Genome Biol Evol 6:1234–1237
Graur D et al (2013) On the immortality of television sets: “function” in the human genome according to the evolution-free gospel of ENCODE. Genome Biol Evol 5:578–590
Dunham I et al (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74
Acknowledgments
I want to thank Christophe Dessimoz, Pascale Gaudet, Jenna Hastings, and Ridhima Kad for helpful discussions on this topic, and for thoughtful reading and suggestions on the manuscript. Open Access charges were funded by the University College London Library, the Swiss Institute of Bioinformatics, the Agassiz Foundation, and the Foundation for the University of Lausanne.
Open Access This chapter is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, duplication, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, a link is provided to the Creative Commons license and any changes made are indicated.
The images or other third party material in this chapter are included in the work’s Creative Commons license, unless indicated otherwise in the credit line; if such material is not included in the work’s Creative Commons license and the respective action is not permitted by statutory regulation, users will need to obtain permission from the license holder to duplicate, adapt or reproduce the material.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
This chapter is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, duplication, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, a link is provided to the Creative Commons license and any changes made are indicated.
The images or other third party material in this chapter are included in the work's Creative Commons license, unless indicated otherwise in the credit line; if such material is not included in the work's Creative Commons license and the respective action is not permitted by statutory regulation, users will need to obtain permission from the license holder to duplicate, adapt or reproduce the material.
Copyright information
© 2017 The Author(s)
About this protocol
Cite this protocol
Thomas, P.D. (2017). The Gene Ontology and the Meaning of Biological Function. In: Dessimoz, C., Škunca, N. (eds) The Gene Ontology Handbook. Methods in Molecular Biology, vol 1446. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-3743-1_2
Download citation
DOI: https://doi.org/10.1007/978-1-4939-3743-1_2
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-3741-7
Online ISBN: 978-1-4939-3743-1
eBook Packages: Springer Protocols