Abstract
This paper focuses on the retrospective evaluation of physiologically based pharmacokinetic (PBPK) techniques used to mechanistically predict clearance throughout pediatric life. An intravenous tramadol retrograde PBPK model was set up in Simcyp® using adult clearance values, qualified for CYP2D6, CYP3A4, CYP2B6, and renal contributions. Subsequently, the model was evaluated for mechanistic prediction of total, CYP2D6-related, and renal clearance predictions in very early life. In two in vitro pediatric human liver microsomal (HLM) batches (1 and 3 months), O-desmethyltramadol and N-desmethyltramadol formation rates were compared with CYP2D6 and CYP3A4 activity, respectively. O-desmethyltramadol formation was mediated only by CYP2D6, while N-desmethyltramadol was mediated in part by CYP3A4. Additionally, the clearance maturation of the PBPK model predictions was compared to two in vivo maturation models (Hill and exponential) based on plasma concentration data, and to clearance estimations from a WinNonlin® fit of plasma concentration and urinary excretion data. Maturation of renal and CYP2D6 clearance is captured well in the PBPK model predictions, but total tramadol clearance is underpredicted. The most pronounced underprediction of total and CYP2D6-mediated clearance was observed in the age range of 2-13 years. In conclusion, the PBPK technique showed to be a powerful mechanistic tool capable of predicting maturation of CYP2D6 and renal tramadol clearance in early infancy, although some underprediction occurs between 2 and 13 years for total and CYP2D6-mediated tramadol clearance.
Similar content being viewed by others
INTRODUCTION
The present study focuses on the mechanistic prediction of tramadol clearance over the pediatric lifespan, since this parameter drives the total exposure to the drug and is key in titrating to the right drug dosage. Physiologically based pharmacokinetic (PBPK) modeling enables pediatric pharmacological research to proceed to the next level, by guiding dose finding studies and the formulation design for the different stages in pediatric life (1–4). The stronghold of PBPK models is that they separate intrinsic from extrinsic patient factors, i.e., drug-independent (system) factors versus drug-dependent factors and study design, so that a physiologically plausible generic human model structure can be applied for any drug under various study conditions (3,5). In addition, a PBPK model, which incorporates demographical, physiological, and biochemical data elements, can be adjusted depending on a specific subpopulation of interest in order to make mechanistic PK predictions. In pediatrics, these models account for the ontogeny of different enzymes, changing tissue volumes and composition, as well as blood flows to these organs (6). At this time, research into the ontogeny of transporters (7,8) lags behind that of hepatic enzymes.
Tramadol is a centrally acting analgesic drug mediating its effect through noradrenaline re-uptake inhibition, increased release of serotonin, and decreased re-uptake of serotonin in the spinal cord. Although in healthy volunteers tramadol is excreted unchanged in urine for about 25%, the greater part is metabolized (9). Tramadol is oxidized to an active metabolite O-desmethyltramadol (ODT) via CYP2D6, having a MU-opioid activity 200 times that of the parent drug. Additionally, tramadol is inactivated to N-desmethyltramadol (NDT) by CYP3A4 and CYP2B6. Secondary metabolism involves further oxidation and phase 2 conjugative reactions (10). In early life, however, these primary metabolic pathways mature differently, and as a consequence, not only the absolute value of the clearance but also relative contributions of the different eliminating pathways may change over time. Therefore, tramadol serves as a model drug in our PBPK study to evaluate whether maturation of the clearance is adequately predicted over the pediatric lifespan. In order to make mechanistically sound pediatric tramadol exposure predictions, different elimination pathways in the tramadol clearance should first be validated in an adult PBPK model previous to predicting pediatric exposure (11). In the application of this methodology, these limitations/assumptions should be considered: (i) clearance pathways in children are the same as those observed in adults, (ii) enzyme kinetics of metabolic contributors are first order, (iii) clearance models are perfusion limited (well-stirred), and (iv) no transporters are involved for which ontogeny information is unknown. These assumptions should always be checked since they are key in the mechanistic prediction of pediatric PK from adult data. Violation of these assumptions may result in clearance under- or overprediction if the clearance pathways differ between children and adults (12), if enzyme processes become saturated or enzyme affinities decrease (e.g., due to the presence of free fatty acids or bilirubin), and if liver/kidney perfusion is altered (e.g., in the case of cirrhosis). Neglecting important transporter involvements might substantially over- or underpredict the clearance, depending on the specific transport mechanism at hand (13). In the case of tramadol, although no OCT1-transport is involved (14), a minor contribution of proton-dependent efflux pumps could not be ruled out (15).
In this work, first, an adequate intravenous adult PBPK model was set up in the Simcyp simulator that accurately describes the total adult clearance, as well as the different CYP450 contributions (CYP2D6, CYP2B6, CYP3A4), and the renal elimination part, composing this clearance. Second, system-specific parameters were adjusted to represent the physiological and biochemical changes occurring during childhood. The mechanistic prediction of the formation clearance to ODT and NDT in pediatrics has to take into account (i) the ontogenic profiles of the CYP isoforms 2D6, 3A4, and 2B6 (16), as well as changes in (ii) the amount of microsomal proteins per gram of liver, (iii) liver size, (iv) liver blood flow, and (v) plasma protein binding (17,18). Third, on the one hand, CYP maturation functions for CYP2D6 (17) and CYP3A4 (18) were compared to experimentally determined human liver microsomal (HLM) activities of tramadol (together with the activity of CYP-specific probe substrates dextromethorphan (DEX for CYP2D6) and midazolam (MDZ for CYP3A4)) in two pediatric batches of 1 and 3 months of age. On the other hand, the pediatric PBPK in vivo clearance predictions were compared to popPK-derived maturation functions (19,20).
MATERIALS AND METHODS
Chemicals and Reagents
All chemicals and reagents used were of the highest available grade: Na2HPO4, KH2PO4, KCl, MgCl2, NADP, HCl (Merck, Darmstadt, Germany), glucose-6-phosphate, glucose-6-phosphate dehydrogenase (Roche Diagnostics GmbH, Mannheim, Germany), tramadol, midazolam (MDZ), dextromethorphan (DEX), O-desmethyltramadol (ODT), N,O-didesmethyltramadol (NODT), 4-OH midazolam, dextrorphan, O-desmethyltramadol-D6, and deuterated alfa-OH midazolam (TRC INC., Toronto, Canada), N-desmethyltramadol (NDT) (LGC GmbH, Luckenwalde, Germany), chlorpropamide (Sigma-Aldrich, St. Louis, USA).
Adult Versus Pediatric In Vitro Metabolism of Tramadol and Probe Substrates MDZ and DEX
Tramadol HLM Incubation Conditions
One pooled adult batch and two pediatric human liver microsomal (HLM) batches (BD Biosciences, Woburn, USA) were used in this study, collected from children aged 1 (male, Caucasian, head trauma) and 3 months (male, Hispanic, anoxia), and stored at −80°C in an Ultra Freezer (New Brunswick scientific, Rotselaar, Belgium). Incubation mixtures (total volume 600 μL) consisted of 297 μL microsomal protein, 3 μL of a tramadol dissolved in MeOH, and 300 μL cofactor mix containing an NADPH-regenerating system consisting of 1 mg of glucose-6-phosphate, 0.50 units of glucose-6-phosphate dehydrogenase, 0.25 mg of NADP, and 1 mg of MgCl2.6H2O in 1 mL of 0.5 M Na,K-phosphate buffer pH 7.4. A preincubation with cofactor mix was done for 5 min in a shaking water bath at 37°C (100 oscillations/min) (Thermo, Waltham, USA). Incubations were started by adding 3 μL of a substrate solution, and stopped by transferring 100 μL aliquots into 96-well plates containing 10 μL ice-cold 4 N HCl and 10 μL of internal standard (O-desmethyltramadol-D6, 6 ng/mL).
Linearity of tramadol metabolite formation was assessed as a function of time and protein concentration at the lowest in vitro substrate concentration in the adult HLM batch. The formation rate was linear up to 10 min and 1 mg protein/mL (data not shown). The enzyme kinetic parameters of tramadol in pediatric HLM were assessed by using a range of incubation concentrations (0.5, 1, 5, 20, 50, 100, 150, 250, 300, and 500 μM). For each substrate and protein concentration level, samples were incubated in duplicate or triplicate and boiled control incubates were run in parallel to correct for non-enzymatic degradation. 96-well plates were then stored at −20°C awaiting to be analyzed by UPLC-MS/MS.
MDZ and DEX HLM Incubation Conditions
MDZ/DEX incubation materials and methods were essentially the same as those for the tramadol metabolism assays. The incubation mixture consisted of 120 μL diluted microsomes, 100 μL cofactor mix for NADPH regeneration system, and 5 μL test compound. After a preincubation period of 5 min at 37°C and 100 oscillations/min, NADP was added to the preincubation mixture to a final volume of 250 μL to initiate the reaction. Midazolam was incubated at 0.2 μM and 0.05 mg protein/mL. The reaction was stopped after 10 min with 250 μL DMSO containing deuterated alfa-OH midazolam as the internal standard (0.1 μg/mL). Dextromethorphan was incubated at 0.5 μM and 0.3 mg protein/mL. The reaction was stopped after 10 min with 250 μL DMSO containing chlorpropamide as the internal standard (0.22 μg/mL). Samples were centrifuged for 10 min at 1711 g and the supernatants introduced to the UPLC-MS system. The intrinsic clearance was calculated in the Enzyme Kinetics module of Sigma Plot.
Bioanalysis
Tramadol’s main metabolites O-desmethyltramadol (ODT, M1), N-desmethyltramadol (NDT, M2), and N,O-didesmethyltramadol (NODT, M5) were quantified by a sensitive UPLC-MS/MS method. A Nexera UHPLC system (Shimadzu, Kyoto, Japan) was coupled to an API 4000 QTRAP (AB Sciex, Toronto, Canada) equipped with a Turbo V™ ion source in ESI+ mode. For the chromatographic separation, a gradient was run—with solvents A (0.025 M ammonium acetate, pH 8.5) and B (acetonitrile:methanol 80:20, v/v)—from 5 to 50% B in 3 min, to 100% B in an immediate step gradient, held for 0.3 min, and back to 5% B, allowing 2 min re-equilibration, at a flow rate of 0.6 mL/min. The column was an Acquity UPLC BEH C18 (1.7 μm) 50 × 2.1 mm column (Waters, Milford, USA), maintained at 60°C. More details about this method were published earlier by our group in T’jollyn et al. (16) and will not be repeated here.
The MDZ metabolite, 4-OH midazolam, was analyzed using a Waters Acquity UPLC system coupled to a Thermo LTQ mass spectrometer (Thermo Fisher Scientific, San Jose, US) in APCI+. The column was an Acquity UPLC BEH C18 (1.7 μm) 50 × 2.1 mm held at 60°C with mobile phase constituents 0.1% HCOOH in ULC water and 0.1% HCOOH in CH3CN in a linear gradient. Run time was 3 min and flow rate 0.6 mL/min. Mass transitions for 4-OH midazolam and deuterated 1-OH midazolam (internal standard) using a collision energy (CE) of 30 eV were 342 > 325, and 346 > 328, respectively. Calibration curves were always made in the same microsomal matrix as the incubates using at least eight calibrator levels and three QC levels for the calibration curve. The DEX metabolite, dextrorphan, was analyzed using a Waters Acquity UPLC system coupled to a Micromass Quattro Ultima triple quadrupole, operating in ESI+. The column was an Acquity UPLC BEH C18 (1.7 μm) 50 × 2.1 mm at 35°C with mobile phase constituents 0.1% HCOOH in ULC water and 0.1% HCOOH in CH3CN in a linear gradient. Run time was 5.25 min and flow rate 0.4 mL/min. Mass transitions for dextrorphan and chlorpropamide using a CE of 28 and 25 eV were 258 > 157, and 277 > 275 (21), respectively.
Enzyme Kinetic Data Analysis
Concentrations of metabolites in the incubation samples were corrected for protein concentration (mg microsomal protein/mL), reaction time (min), and initial substrate concentration (μM) in order to calculate the apparent in vitro clearance (CLapp) for every metabolite. For tramadol, CLapp was plotted versus tramadol incubation concentration and a nonlinear model—with the model structure provided in Eq. 1—was fitted to the data, using R v3.1.1 (22). Models were evaluated by visually inspecting residual plots for bias. In Eq. 1, CL app is the apparent in vitro clearance, v o is the initial rate of metabolite formation in the incubate, [S] is the tramadol concentration (μM), Km is the Michaelis-Menten constant (μM), and Vmax is the maximum velocity. This equation allowed estimation of the parameters Km and Vmax, and hence the calculation of CLint. An unbound fraction in microsomes (fumic) of ~0.96 was estimated in silico using the prediction toolbox in Simcyp® v13 (23).
Development of the Adult Tramadol PBPK Model
An intravenous adult PBPK model was set up, using the retrograde calculator available in the Simcyp Simulator (v13, Sheffield, UK), by extracting in vivo hepatic and renal clearance values from publications available in the scientific literature (24–28). In this retrograde calculator, hepatic intrinsic clearances per CYP isoform are calculated, based on hepatic plasma clearance and apparent in vivo CYP contributions, using the well-stirred liver model (29,30). Table S1 with final numerical values for different parameters is provided in the supplementary data. The following approaches were used to define the contribution of CYP2D6, CYP2B6, and CYP3A4 in the adult hepatic clearance of tramadol.
CYP2D6 Contribution
The CYP2D6 contribution in the tramadol hepatic clearance was assessed by comparing the hepatic clearance increase between poor and extensive metabolizers in observations and predictions. Observations were extracted from a study conducted by Pedersen et al. (31). Patients from this study were genotyped as *1/*1 (EM; n = 8) and *4/*4 (n = 7) or *4/*6 (PM) (n = 1). By taking into account the actual age range, administered dose, and proportion of males and females in the PBPK trial design, we mimicked the actual clinical trial in our predictions. Virtual populations are generated in the Simcyp Simulator by using a correlated Monte Carlo approach. For a more detailed description, we redirect the interested reader to Jamei et al. (32).
CYP2B6-CYP3A4 Contribution
Although CYP2B6 seems to be a minor contributor to the total clearance, the CYP2B6 contribution apart from the CYP3A4 involvement was assessed by performing a tramadol-rifampicin drug-drug interaction (DDI) simulation and by comparison of predicted with observed area under the curves (AUCs). Using information of the study population and design by Saarikoski et al. (33), 100 virtual trials were simulated with 12 subjects each receiving 6 doses of 600 mg oral rifampicin (RIF) every 24 h were simulated. Twelve hours after the last rifampicin dose, 50 mg tramadol was administered and the change in AUC reported. Subjects ranged from 18 to 30 years of age, and the female proportion was set to 0.42. Drug-specific parameters for rifampicin (elimination and induction effects on CYP2B6 and CYP3A4), described elsewhere (34), were used in this simulation. While keeping the CYP2D6 contribution in the tramadol adult PBPK model fixed, the CYP2B6 contribution was increased from 0 to 30% while the CYP3A4 contribution was (in parallel) decreased from 52 to 22%. The sum of the contributions of CYP2D6 and CYP2B6-3A4 always added up to 100% of the hepatic intrinsic clearance. Results for the 100 simulated trials were expressed as the geometric mean ratio of the AUCcontrol/AUCinduced and were compared to the observed geometric mean ratio from the actual in vivo DDI study (33).
Prediction of Pediatric Clearance with the Developed Tramadol PBPK Model
The developed PBPK model was used to predict the clearance (total, CYP2D6, and renal component) over the pediatric lifespan, using the pediatric module of the Simcyp® Simulator. This module contains ontogeny information for the different CYP450 enzymes considered in this work (CYP2D6 and CY2B6 ontogeny functions are described by Johnson et al. in (17), whereas CYP3A4 ontogeny information is described by Salem et al. in (35)). Additionally, the changes in tissue volumes (i.e., liver, kidney), tissue composition (lipids vs. water content), and blood flows (i.e., QH, GFR), also described in (17), all vary as a function of age and determine the extent of the clearance in a specific pediatric subject.
Model Evaluation
On the in vitro level, theoretical CYP maturation functions for CYP2D6 (17) and CYP3A4 (18) were compared to experimentally determined human liver microsomal (HLM) activities of tramadol (together with the activity of CYP-specific probe substrates dextromethorphan (DEX for CYP2D6) and midazolam (MDZ for CYP3A4)) in two pediatric batches of 1 and 3 months of age. On the in vivo level, pediatric PBPK model predictions (from a full term 40 weeks PMA onwards) were visually compared to in vivo clearance maturation models, in terms of total, CYP2D6, and renal tramadol clearance, published in literature and described in more detail in the “Available Pediatric In Vivo Reference Data” section.
Available Pediatric In Vivo Reference Data
Clearance maturation parameter estimates were collected from publications in the scientific literature (19,20). The overall maturation function (maturation + size function) was applied to predict individual clearance values for extensive CYP2D6 metabolizers only, given the subjects’ age (post-menstrual age; PMA) and weight (kg). Besides, the raw clinical data of a subset of 57 neonates and young infants, used for estimating total, CYP2D6 and renal clearance parameters, were available.
Exponential Maturation Function
The first published maturation function for total and CYP2D6-mediated tramadol clearance originates from a publication by Allegaert et al. (20). This resulting maturation curve for the total clearance is obtained by summation of “CYP2D6” and “non-CYP2D6” clearance maturation, both taking on following form:
in which CLstd is the standardized clearance, Fsize is the factor taking into account the size effect (WTi/WTstd)**0.75, and SLPCL (slope of clearance) is the exponent of the maturation function centered around a term age of 40 weeks PMA. Numerical values for these parameters are provided in Table S2 in the supplementary data.
Hill Maturation Function
The second maturation function was derived from a pooled popPK analysis (19) in pediatric and adult subjects in which the maturation of “CYP2D6” and “non-CYP2D6” tramadol clearance was described by a Hill function, as follows:
in which TM50 is the age (PMA) at 50% of maturation, and Hill is the Hill coefficient describing the steepness of the maturation function. Numerical values for these parameters are provided in Table S2 in the supplementary data. Both modeling attempts assume a two-compartmental, linear disposition model for tramadol with first-order elimination, consisting of different clearance pathways, based on plasma observations of parent tramadol and ODT metabolite. The major difference between these two models lies in how the PK of the metabolite is described. In the first model, the metabolite volume is fixed to 224 L/70 kg and metabolite clearance is estimated to exponentially increase with PMA (20). In the second model, the metabolite volume is estimated by applying allometric principles to an observed metabolite volume in dogs. Metabolite clearance is fixed by a Hill function assuming a maturation of the glomerular filtration rate (19).
WinNonlin® Fitting of Subjects with Rich Sampling
In addition to the previously described popPK models, which are solely based on plasma observations, we performed a one-by-one fitting procedure in WinNonlin®, taking into account not only plasma observations but also urinary excretion data of parent and metabolite (36). These were available for 9 out of 57 neonates and young infants from the original dataset, which were richly sampled. The compartmental model depicted in Fig. 1 was used to estimate the different clearance parameters. Pharmacokinetic analysis for the one-by-one fitting was performed by WinNonlin Professional version 5.2.1. (Pharsight, St-Louis, MI, USA). Plasma and urinary concentrations of parent and metabolite were simultaneously modeled, applying a user-written differential equation model. Both plasma and urine concentrations were modeled using the 1/Y weight, and the Gauss-Newton (Levenberg and Hartley) algorithm with 50 iterations. A one-compartment model for tramadol was retained on the basis of visual inspection of the fitted plasma concentration-time profiles and standard errors of the estimated PK parameters. Tramadol was cleared by renal excretion (CLPR), CYP2D6 clearance (CLP2M), and other clearance routes (CLPO). For the CYP2D6 metabolite, ODT, a one-compartment model was used with formation clearance (CLP2M), ODT moiety-related clearance (CLM), and volume (VM). The metabolite clearance (CLM) is a hybrid clearance parameter describing the total mass outflux of ODT, since ODT concentrations measured in urine are the sum of unchanged as well as conjugated ODT (36).
A one-compartment linear disposition model was used to fit the parent drug. An additional compartment for the ODT metabolite was linked to the central compartment by its formation clearance (CLP2M). Urinary excretion of tramadol (CLPR) was estimated based on the total amount of tramadol found in urine (UR). ODT moiety-related clearance (CLM) and volume (VM) were estimated based on the ODT plasma concentrations and total ODT amount found in urine (UR); CLPO other clearance parent
RESULTS
Qualification of Tramadol Adult PBPK Model
By scaling back intravenous tramadol in vivo adult clearance data, the tramadol adult PBPK model is provided with information on the CYP2D6 involvement by comparing hepatic clearances between different CYP2D6 metabolizer statuses (31), and the CYP2B6-CYP3A4 involvement by comparing AUC ratios after induction of tramadol metabolism by rifampicin (33) (Table I). CYP2D6 was calculated to be involved for 48% in the hepatic clearance, based on an in vivo observed 1.74-fold hepatic clearance increase from CYP2D6 poor metabolizer (PM, no remaining CYP2D6 activity) to extensive metabolizers (EM). The DDI clinical trial simulation of tramadol and rifampicin was used to calculate the relative CYP2B6-3A4 involvement. The observed geometric mean AUC ratio (induced/control) fell within the 90% confidence interval around the predicted geometric mean AUC ratio, based on 100 virtual trials mimicking the actual in vivo trial (Fig. 2), only if the CYP2B6 contribution in the hepatic intrinsic clearance was less than 10% and the CYP3A4 contribution not below 42%. Therefore, the percentage CYP2B6 involvement in hepatic tramadol metabolism was estimated to be not more than 10%. As a consequence, the CYP3A4 contribution was estimated to constitute between 42 and 52% of tramadol hepatic metabolism (Table I).
DDI clinical trial simulation of tramadol and rifampicin to assess the CYP2B6-3A4 contribution. The figure depicts the results of 100 trial simulations mimicking the original trial, when CYP2B6-3A4 contributions are assumed to be 10–42%, respectively. Black horizontal lines represent the in vivo observed ratio of AUC geo means (solid) and 90% confidence limits (dashed). The solid red line represents the average ratio of simulated AUC geo means. Since the solid black line still is in the 90% confidence region (grayed area) of the simulated ratio, we assume that CYP2B6 contribution should be between 0 and 10%
Adult Versus Pediatric In Vitro Metabolism of Tramadol and Probe Substrates MDZ and DEX
The in vitro metabolism of tramadol to its two primary metabolites (ODT and NDT) is displayed for pediatric HLM batches of 1 and 3 months of age in Figs. S1 and S2, respectively, provided in the supplementary data. In vitro clearance values were modeled versus the tramadol incubation concentration. Additionally, fractional activities were measured in both pediatric HLM batches versus the adult HLM pool for tramadol’s primary metabolites, as well as for the probe substrates dextromethorphan (DEX) and midazolam (MDZ), providing information about the activity of CYP2D6 and CYP3A4 in these batches, respectively. Fractional activities for either ODT/NDT, DEX, or MDZ (Table II) were calculated as the ratio of their intrinsic clearances, i.e., CLintpedHLM/CLintadultHLM.
The lines in Fig. 3, panel A display the “assumed” or “theoretical” maturation of CYP2D6 (17) and CYP3A4 (18) on the per mg microsomal protein level, expressed as fractional activity relative to adult up to 1 year of age, whereas the points display in vitro measured fractional activities for the two pediatric batches, which are also displayed in Table II. These values represent measured fractional activities of the pediatric intrinsic clearance relative to the measured adult intrinsic clearance for tramadol, midazolam (MDZ), and dextromethorphan (DEX). MDZ and DEX were included as probe substrates in the in vitro HLM batches in order to correct for batch-specific activities of CYP3A4 and CYP2D6, respectively. Fractional activity of ODT formation in the pediatric batches is 1.6- and 1.9-fold higher than in the pooled adult HLM batch, for 1 and 3 months of age, respectively. This is highly analogous to the fractional activity of DEX for the two pediatric batches, being 1.6- and 1.7-fold higher (Table II). Correction of the measured batch-specific fractional activities for ODT and NDT formation with the theoretical fractional activities of DEX and MDZ, denoted by Eq. 4, can be found in Fig. 3b. Because the corrected fractional activity of ODT formation for both batches (“corrODT 1 M” and “corrODT 3 M”) strictly follows the maturation profile of CYP2D6, ODT formation is highly correlated with CYP2D6 maturation.
Theoretical maturation of CYP2D6 (17) (red line) and CYP3A4 (18) (black line) expressed as fractional activity versus age. In the left plot, ODT/NDT activity (solid dots) for two pediatric HLM batches (1 and 3 months) are depicted together with the activity of probes (open triangles) DEX (act2D6) and MDZ (act3A4). In the right plot, the ratio of each corresponding pair is taken as described in the body text and indicated as corrODT and corrNDT. ODT formation follows the CYP2D6 maturation profile, while NDT formation does not follow the CYP3A4 maturation profile
in which corrODT is the fractional activity of ODT formation corrected by the batch-specific fractional activity of DEX at a given age i, fractODT is the fractional activity of ODT formation for a given age i, and d(fractDEX-theorDEX) is the distance from the measured fractional activity of DEX to the theoretical maturation of DEX at a given age i (i.e., 1 and 3 months) (analogous for NDT).
Tramadol’s NDT formation was not well correlated with MDZ metabolism, as the NDT formation corrected by the batch-specific CYP3A4 activity for the 3 months batch is not following the maturation profile in Fig. 3b.
Comparison of Pediatric PBPK Predictions with Pediatric In Vivo Reference Data
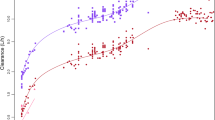
Pediatric PBPK predictions of total tramadol clearances were compared to tramadol clearances derived using two published maturation models (exponential and Hill-type), as well as from a WinNonlin® fit incorporating urine observations. Figure 4 displays the total tramadol clearance as a function of PMA (left) and body weight (right). The Hill and exponential model are depicted by a solid black and red line, respectively, while the WinNonlin® fit is illustrated as light brown points together with their smoother function. Dark blue points are the mechanistic pediatric PBPK predictions of the total tramadol clearance. An underprediction of the PBPK-predicted total tramadol clearance is apparent from both plots (i.e., as a function of PMA or weight) when comparing with any of the in vivo maturation models.
Figure 5 displays the maturation of tramadol clearance mediated by CYP2D6 as a function of PMA (left) and bodyweight (right). Noticeable from these plots is that PBPK predictions of tramadol CYP2D6 clearance are in line with the Hill model (black line) and the WinNonlin® fits, but not with the exponential maturation model.
Figure 6 depicts the maturation of renal clearance (as a function of PMA and bodyweight) and enables comparison of PBPK-predicted renal clearance and in vivo observed renal clearance. In vivo estimates for the renal clearance of unchanged tramadol are only available from the WinNonlin® fits because only in this modeling approach, urinary data was considered. PBPK-predicted renal clearance is very analogous to the in vivo estimates.
Finally, Figs. 7 and 8 display the complete age span from 40 to 1300 weeks PMA (25 years) for the total and CYP2D6 clearance maturation, comparing PBPK predictions (blue dots) and Hill function predictions (red dots). Tramadol clearance is underpredicted over the complete pediatric life span. CYP2D6 clearance predictions are in line with in vivo observations in very early life (40–53 weeks PMA, Fig. 8), but start to deviate from the in vivo maturation trend from 53 weeks PMA (1 year) onwards until 700 weeks PMA (13 years). For adults, predictions are again in line with in vivo.
DISCUSSION
The first step in building a pediatric PBPK model is to develop an accurate, robust adult PBPK model. Although our group already demonstrated that PBPK models of tramadol could be built up from actual HLM or rhCYP in vitro enzyme kinetic data (16), we choose to use a PBPK modeling approach that was built from in vivo clearance values, to be able to predict pediatric PK, since it is the most accurate model for describing adult tramadol PK in healthy volunteers. In this approach, intrinsic clearance values are calculated from in vivo clearance values from healthy adult volunteers, using a well-stirred model, to obtain a PBPK model that accurately predicts the in vivo clearance. In addition, specific CYP450 enzyme contributions can be incorporated in this adult PBPK model, which is described in the “MATERIALS AND METHODS” section. Pediatric predictions derived from this adult PBPK model are mechanistic because all relevant pathways observed in adults are used for predicting the PK in pediatrics, based on knowledge of functionality maturation in different tissues (e.g., liver eliminating capacity, renal filtration rate, etc.). Although in the adult PBPK model a CYP2D6, a CYP2B6-CYP3A4, and a renal component were implemented, we only focused on prediction of the total, CYP2D6 and renal clearance component for the pediatric populations, since in vivo reference data were available for these pathways. After having studied the adult in vitro metabolism in HLM, we assessed whether formation of the primary metabolites O-desmethyltramadol (ODT) and N-desmethyltramadol (NDT) in pediatric HLM is governed by the same enzymes as in adult HLM. The result of this experiment would indicate if our approach of applying known maturation functions (17,35) of the responsible CYP450 enzymes in adults to predict the pediatric clearance is valid. Therefore, in order to assess the fractional activity of ODT and NDT formation relative to adult HLM, pooled adult HLM were incubated with tramadol, as well as probe substrates DEX (CYP2D6) and MDZ (CYP3A4) in pediatric and adult HLM. Fractional activity is calculated by taking the ratio of the activity observed in pediatric HLM and the activity observed in pooled adult HLM, as described in the “MATERIALS AND METHODS” section. High fractional activities were found for ODT (1.6- and 1.9-fold, Table II) which can be explained by the fact that we have observed a relatively low CYP2D6 activity in the pooled adult HLM batch (data not shown). Because we also measured CYP2D6 fractional activity for the probe substrate (Table II), we were able to calculate the difference between the measured CYP2D6 (DEX) fractional activity and the theoretical (based on in vitro ontogeny) CYP2D6 activity. This difference illustrates to what extent the measured CYP2D6 activity deviates from what one would expect for a given age. Correction of the measured ODT fractional activity with the previously calculated difference, which yields “corrODT 1 M” and “corrODT 3 M” in Fig. 3b, confirms us that ODT formation in pediatrics, just as in adults, is CYP2D6-mediated. Applying an analogous correction to NDT formation with measured MDZ activity yields predictions in Fig. 3b that are not in line with the CYP3A4 activity at all. A possible explanation is that CYP3A4 is not the only CYP enzyme contributing to the formation of NDT, also confirmed by other reports (16,37). This comparison at the in vitro level enabled us to validate the assumption that in adult as well as in early life, CYP2D6 is the major isoform contributing to the metabolism of tramadol to O-desmethyltramadol, making this metabolite a valid probe to study CYP2D6 maturation. Nevertheless, it would be valuable to test more pediatric batches in addition to the pediatric batches described in this study in order to compare/validate the ontogeny functions for the enzymes of interest over a broader age range. In addition, since the ontogeny data for different CYP450 isoforms are available, the extent of pediatric in vitro metabolism can be predicted, extrapolating from adult in vitro metabolism data. This approach can serve as guidance for interpreting pediatric metabolism studies, e.g., a significant elevation of an assumed 3A4 metabolite in combination with an anticipated low 3A4 activity, e.g., may indicate CYP3A7 involvement, if substrate overlap is expected.
Pediatric PBPK predictions of tramadol were performed with the pediatric module of the Simcyp® Simulator, which was first used for the setup of an adult PBPK model. At the in vivo level, the maturation of total tramadol clearance could be compared between PBPK-predicted and in vivo observed data sets (Fig. 4). The clearance as a function of weight or PMA by the PBPK model is underpredicted compared to the in vivo maturation profiles. As has been described above, the clearance prediction for this PBPK model is based on three hepatic components (CYP2D6, CYP3A4, and CYP2B6) and one renal component (GFR). Possible explanations for an underprediction are (i) incorrect maturation function for either of the implemented CYP enzymes, (ii) other metabolic pathways contributing to tramadol’s metabolism in pediatric life (FMO, CYP3A7) (12,38), or (iii) neglecting other minor pathways in tramadol metabolism (10). Therefore, the total clearance of tramadol was decomposed in its constituents and compared to in vivo observations, when possible. To this end, (i) since we had strong in vitro proof that ODT is a good CYP2D6 activity probe, we compared the predicted versus in vivo observed tramadol CYP2D6 clearance increase, documented in Fig. 5, and (ii) we compared the tramadol renal clearance increase versus in vivo maturation, illustrated in Fig. 6. Concerning the renal clearance maturation, we could only compare our PBPK predictions to the WinNonlin® fits (light brown) since this was the only in vivo model that incorporated urinary data for estimation of the clearance parameters. The comparison of the renal clearance, plotted as a function of PMA, between PBPK and WinNonlin® fits yielded good agreement, while the comparison as a function of WT was less good but still acceptable. As for the CYP2D6 clearance maturation, PBPK predictions are in close agreement with the Hill maturation model (black) and the WinNonlin® fits (light brown), but not with the exponential maturation model (red). So which in vivo model is the best representation of the CYP2D6 clearance increase? Both the exponential and the Hill model are population PK models built on plasma observations for parent and ODT metabolite, and share an allometric relationship in their covariate structure, but the age models are respectively exponential and Hill-type. Furthermore, the exponential model is based on plasma observations for 57 neonates and young infants, using a fixed metabolite volume of 224 L/70 kg, whereas the Hill model incorporates plasma observations from 295 subjects from early to adult life and estimates a metabolite volume based allometric principles from dogs. Because in neither model urinary data was included, we performed a WinNonlin® fitting procedure including urinary data of parent and metabolite. This allowed estimation of the ODT moiety-related volume and its associated clearance value, as well as volume and clearance of tramadol. Although the WinNonlin® parameter estimates are few in number, they agree with the Hill function as descriptor for maturation in CYP2D6 clearance. We believe that the Hill model is the better representation of clearance maturation in pediatrics because this maturation function is physiologically more plausible, since the “developmental” processes of cells tend to reach their end state at a certain point in childhood, while “growth” processes take over the increasing elimination capacity of such organs. As the Hill model is also based on the larger data set, we choose the Hill model as a more correct in vivo model to compare the PBPK predictions against. It turns out that the CYP2D6 clearance maturation from PBPK aligns much better with our WinNonlin® fits and the Hill model than with the exponential model. Finally, the maturation from birth to adult life is presented for total and CYP2D6 tramadol clearance in Figs. 7 and 8, respectively. The maturation increase for the total and the CYP2D6 clearance seems to agree with the in vivo model in very early life, with CYP2D6 absolute clearance values that are also in line with in vivo observed values. However, until the age of 13 (700 weeks PMA), both predicted clearance parameters display a relatively constant underprediction, which disappears again in adulthood. It might be that in children, the maturation of liver size as a function of bodyweight is underestimated, since demographic parameters in Simcyp® are mainly based on UK growth charts (personal communication). We acknowledge that choosing the ages where clearances deviate significantly from predictions can be subject to discussion, which again highlights the need for developing adequate evaluation techniques in PBPK.
CONCLUSION
In conclusion, PBPK modeling and simulation techniques allow a mechanistic prediction of maturation of CYP2D6-mediated and renal tramadol clearance in very early life. Some underprediction occurred between 2 and 13 years for total and CYP2D6 tramadol clearance. Care has to be taken when selecting in vivo maturation models as a reference, since these “top-down” models also rely on assumptions made.
References
Maharaj AR, Barrett JS, Edginton AN. A workflow example of PBPK modeling to support pediatric research and development: case study with lorazepam. Aaps J. 2013;15(2):455–64.
Manolis E, Osman TE, Herold R, Koenig F, Tomasi P, Vamvakas S, et al. Role of modeling and simulation in pediatric investigation plans. Paediatr Anaesth. 2011;21(3):214–21.
Zhao P, Zhang L, Grillo JA, Liu Q, Bullock JM, Moon YJ, et al. Applications of physiologically based pharmacokinetic (PBPK) modeling and simulation during regulatory review. Clin Pharmacol Ther. 2011;89(2):259–67.
Bellanti F, Della PO. Modelling and simulation as research tools in paediatric drug development. Eur J Clin Pharmacol. 2011;67 Suppl 1:75–86.
Jamei M, Dickinson GL, Rostami-Hodjegan A. A framework for assessing inter-individual variability in pharmacokinetics using virtual human populations and integrating general knowledge of physical chemistry, biology, anatomy, physiology and genetics: a tale of ‘bottom-up’ vs ‘top-down’ recognition of covariates. Drug Metab Pharmacokinet. 2009;24(1):53–75.
Johnson TN, Rostami-Hodjegan A. Resurgence in the use of physiologically based pharmacokinetic models in pediatric clinical pharmacology: parallel shift in incorporating the knowledge of biological elements and increased applicability to drug development and clinical practice. Pediatr Anesth. 2010;21(3):291–301.
Mooij MG, Schwarz UI, de Koning BA, Leeder JS, Gaedigk R, Samsom JN, et al. Ontogeny of human hepatic and intestinal transporter gene expression during childhood: age matters. Drug Metab Dispos: Biol Fate Chem. 2014;42(8):1268–74.
Sweet DH, Bush KT, Nigam SK. The organic anion transporter family: from physiology to ontogeny and the clinic. Am J Physiol Renal Physiol. 2001;281(2):F197–205.
Grond S. Clinical pharmacology of tramadol. Clin Pharmacokinet. 2004;43(13):879–923.
Wu WN, Mckown LA, Liao S. Metabolism of the analgesic drug ULTRAM (R) (tramadol hydrochloride) in humans: API-MS and MS/MS characterization of metabolites. Xenobiotica. 2002;32(5):411–25.
Leong R, Vieira ML, Zhao P, Mulugeta Y, Lee CS, Huang SM, et al. Regulatory experience with physiologically based pharmacokinetic modeling for pediatric drug trials. Clin Pharmacol Ther. 2012;91(5):926–31.
Yanni SB, Annaert PP, Augustijns P, Ibrahim JG, Benjamin DK, Thakker DR. In vitro hepatic metabolism explains higher clearance of voriconazole in children versus adults: role of CYP2C19 and flavin-containing monooxygenase 3. Drug Metab Dispos. 2010;38(1):25–31.
Shitara Y, Horie T, Sugiyama Y. Transporters as a determinant of drug clearance and tissue distribution. Eur J Pharm Sci. 2006;27(5):425–46.
Tzvetkov MV, Saadatmand AR, Lotsch J, Tegeder I, Stingl JC, Brockmoller J. Genetically polymorphic OCT1: another piece in the puzzle of the variable pharmacokinetics and pharmacodynamics of the opioidergic drug tramadol. Clin Pharmacol Ther. 2011;90(1):143–50.
Kanaan M, Daali Y, Dayer P, Desmeules J. Uptake/efflux transport of tramadol enantiomers and O-desmethyl-tramadol: focus on P-glycoprotein. Basic Clin Pharmacol Toxicol. 2009;105(3):199–206.
T'Jollyn H, Snoeys J, Colin P, Van Bocxlaer J, Annaert P, Cuyckens F, et al. Physiology-based IVIVE predictions of tramadol from in vitro metabolism data. Pharm Res. 2015;32(1):260–74.
Johnson TN, Rostami-Hodjegan A, Tucker GT. Prediction of the clearance of eleven drugs and associated variability in neonates. Infants Child Clin Pharmacokinet. 2006;45(9):931–56.
Salem F, Johnson TN, Barter ZE, Leeder JS, Rostami-Hodjegan A. Age related changes in fractional elimination pathways for drugs: assessing the impact of variable ontogeny on metabolic drug-drug interactions. J Clin Pharmacol. 2013;53(8):857–65.
Allegaert K, Holford N, Anderson BJ, Holford S, Stuber F, Rochette A, et al. Tramadol and o-desmethyl tramadol clearance maturation and disposition in humans: a pooled pharmacokinetic study. Clin Pharmacokinet. 2015;54(2):167–78.
Allegaert K, van den Anker J, de Hoon J, van Schaik R, Debeer A, Tibboel D, et al. Covariates of tramadol disposition in the first months of life. Br J Anaesth. 2008;100(4):525–32.
De Bock L, Boussery K, Colin P, De Smet J, T'Jollyn H, Van Bocxlaer J. Development and validation of a fast and sensitive UPLC-MS/MS method for the quantification of six probe metabolites for the in vitro determination of cytochrome P450 activity. Talanta. 2012;89:209–16.
Team RC. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2013.
Turner DB, Yeo KR, Tucker GT, Rostami-Hodjegan A. Prediction of nonspecific hepatic microsomal binding from readily available physicochemical properties. Drug Metab Rev. 2006;38:162.
Garcia-Quetglas E, Azanza JR, Cardenas E, Sadaba B, Campanero MA. Stereoselective pharmacokinetic analysis of tramadol and its main phase I metabolites in healthy subjects after intravenous and oral administration of racemic tramadol. Biopharm Drug Dispos. 2007;28(1):19–33.
Lintz W, Barth H, Becker R, Frankus E, Schmidt-Bothelt E. Pharmacokinetics of tramadol and bioavailability of enteral tramadol formulations - 2nd communication: Drops with ethanol. Arzneimittelforschung. 1998;48(5):436–45.
Lintz W, Barth H, Osterloh G, Schmidt-Bothelt E. Pharmacokinetics of tramadol and bioavailability of enteral tramadol formulations - 3rd communication: Suppositories. Arzneimittelforschung. 1998;48(9):889–99.
Lintz W, Becker R, Gerloff J, Terlinden R. Pharmacokinetics of tramadol and bioavailability of enteral tramadol formulations - 4th Communication: Drops (without ethanol). Arzneimittelforschung. 2000;50(2):99–108.
Lintz W, Erlacin S, Frankus E, Uragg H. Metabolismus von tramadol bei mensch und tier. Arzneimittelforschung. 1981;31(11):1932.
Bjorkman S. Prediction of drug disposition in infants and children by means of physiologically based pharmacokinetic (PBPK) modelling: theophylline and midazolam as model drugs. Br J Clin Pharmacol. 2005;59(6):691–704.
Jamei M, Bajot F, Neuhoff S, Barter Z, Yang J, Rostami-Hodjegan A, et al. A mechanistic framework for in vitro-in vivo extrapolation of liver membrane transporters: prediction of drug-drug interaction between rosuvastatin and cyclosporine. Clin Pharmacokinet. 2014;53(1):73–87.
Pedersen RS, Damkier P, Brosen K. Enantioselective pharmacokinetics of tramadol in CYP2D6 extensive and poor metabolizers. Eur J Clin Pharmacol. 2006;62(7):513–21.
Jamei M, Marciniak S, Feng K, Barnett A, Tucker G, Rostami-Hodjegan A. The Simcyp population-based ADME simulator. Expert Opin Drug Metab Toxicol. 2009;5(2):211–23.
Saarikoski T, Saari TI, Hagelberg NM, Neuvonen M, Neuvonen PJ, Scheinin M, et al. Rifampicin markedly decreases the exposure to oral and intravenous tramadol. Eur J Clin Pharmacol. 2013;69(6):1293–301.
Rekic D, Roshammar D, Mukonzo J, Ashton M. In silico prediction of efavirenz and rifampicin drug-drug interaction considering weight and CYP2B6 phenotype. Br J Clin Pharmacol. 2011;71(4):536–43.
Salem F, Johnson TN, Abduljalil K, Tucker GT, Rostami-Hodjegan A. A re-evaluation and validation of ontogeny functions for cytochrome P450 1A2 and 3A4 based on in vivo data. Clin Pharmacokinet. 2014;53(7):625–36.
Allegaert K, van den Anker J, Verbesselt R, de Hoon J, Vanhole C, Tibboel D, et al. O-demethylation of tramadol in the first months of life. Eur J Clin Pharmacol. 2005;61(11):837–42.
Subrahmanyam V, Renwick AB, Walters DG, Young PJ, Price RJ, Tonelli AP, et al. Identification of cytochrome P-450 Isoforms responsible for cis-tramadol metabolism in human liver microsomes. Drug Metab Dispos. 2001;29(8):1146–55.
Ince I, Knibbe CA, Danhof M, de Wildt SN. Developmental changes in the expression and function of cytochrome P450 3A isoforms: evidence from in vitro and in vivo investigations. Clin Pharmacokinet. 2013;52(5):333–45.
Acknowledgments
This study was supported by the “Agency for Innovation by Science and Technology in Flanders (IWT)” through the ‘SAFEPEDRUG’ project (IWT/SBO 130033).
The clinical research of Karel Allegaert is supported by the Fund for Scientific Research, Flanders (Fundamental Clinical Investigatorship 1800214N).
Author information
Authors and Affiliations
Corresponding author
Additional information
Guest Editors: Bernd Meibohm, Jeffrey S. Barrett, and Gregory Knipp
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 200 kb)
Rights and permissions
About this article
Cite this article
T’jollyn, H., Snoeys, J., Vermeulen, A. et al. Physiologically Based Pharmacokinetic Predictions of Tramadol Exposure Throughout Pediatric Life: an Analysis of the Different Clearance Contributors with Emphasis on CYP2D6 Maturation. AAPS J 17, 1376–1387 (2015). https://doi.org/10.1208/s12248-015-9803-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1208/s12248-015-9803-z