Abstract
The TET dioxygenases, TET1, TET2, and TET3, catalyze transfer of an oxygen atom to the methyl group of 5-methylcytocine (5-mC), converting it to 5-hydroxymethylcytocine (5-hmC). Among the genes encoding these enzymes, ten-eleven translocation 2 (TET2) is frequently mutated somatically in both myeloid and lymphoid malignancies. Because these TET2 mutations result in the impairment of the dioxygenase activity of TET2, it is thought that these mutations interfere with 5-mC to 5-hmC conversion. There is ample evidence indicating that TET2 mutations are a driver of tumorigenesis in blood cells and that TET2 mutations are often acquired at the hematopoietic stem/early progenitor cell stage. In addition, TET2 is the second-most frequently mutated gene in clonal hematopoiesis in individuals with no apparent blood cancers, suggesting that while TET2 mutations alone are insufficient to cause hematologic malignancy, they represent an early event during tumorigenesis. A number of questions, including the precise target genome regions of TET2, and the importance of the balance of 5-mC and 5-hmC in the regulatory regions in transcriptional control, remain.
Similar content being viewed by others
Introduction
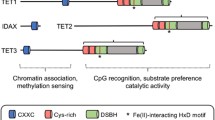
The mechanism by which methylated DNA [particularly 5-methylcytosine (5-mC) at the CpG dinucleotide] is actively demethylated was for many years a central question in the quest to better understand epigenetic regulation. Until recently, however, it remained unsolved, with a history of failures in the discovery of DNA demethylases in animal cells [1]. In 2009, the ten-eleven translocation 1 (TET1) gene was shown to encode a dioxygenase that converts 5-mC to 5-hydroxymethylcytocine (5-hmC) by transferring an oxygen atom to the methyl group of 5-mC [2]. This discovery triggered the subsequent rapid progress in understanding the mechanisms that underlie demethylation processes [3, 4]. TET2 and TET3 were soon found to be products of genes belonging to the same family as TET1 and to exhibit similar dioxygenase functions [5, 6]. Once a TET dioxygenase catalyzes the 5-mC to 5-hmC conversion, multistep biochemical reactions follow, and the position is ultimately replaced by the unmodified cytosine (uC) (Fig. 1).
TET enzymes. These convert 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC). 5hmC is further oxidized to 5-formyl cytosine (5-fC) and 5-carboxyl cytosine (5caC), which enter the demethylation pathway by thymine DNA glycosylase (TDG) and base-excision repair (BER). 5hmC also functions as an epigenetic mark. TET2 lacks the CXXC consensus domain; it is presumed that the N-terminal region of TET2 is expressed as a separate protein, IDAX, by the ancestral chromosomal inversion
Somatic TET2 mutations are frequently identified in a wide variety of hematologic malignancies [7, 8]. These mutations cause impairment of enzymatic activity of the TET2 dioxygenase, resulting in the failure of 5-mC to 5-hmC conversion, and eventually failure of demethylation. The abnormal 5-mC/5-hmC/uC pattern in the TET2 mutation-carrying cells is thought to cause changes in the expression of the target genes and eventually drive the cells to develop hematologic malignancies. However, it remains unclear how TET2 mutations change the scheme of methyl group modification of the cytosine at the CpG sites throughout the genome and how such structural changes affect gene expression profiles.
There have, however, been important advances in our understanding of why TET2 mutations are identified so commonly in such a wide variety of hematologic malignancies and the biological significance of these mutations. In this PIH article, I will focus mainly on the biological aspects of TET2 mutations, followed by a brief review of recent progress in our understanding of their epigenetic aspects.
Frequent somatic TET2 mutations in hematologic malignancies
Frequent somatic mutations in TET2 gene were first discovered in 2009 in myeloid malignancies, such as myelodysplastic syndrome (MDS), chronic myelomonocytic leukemia (CMML), acute myeloid leukemia (AML), and myeloproliferative neoplasms (MPN) [9–12]. Of these myeloid malignancies, TET2 mutations have been found to be the most frequent in CMML [7]. In MDS, TET2 is the most frequently mutated gene [13]. In AML, TET2 mutations are more frequent in secondary than in de novo cases [14] (Fig. 2). These mutations are frame-shift mutations and nonsense mutations throughout the coding region or missense mutations localized to the functional domains required for the enzymatic activity. The crystal structure of the TET2-DNA complex reveals that the region flanking missense mutations is actually important for the catalytic activity of TET2 [15].
Frequencies of TET2 gene mutations in various blood cancers. AML acute myelogenous leukemia, MDS, myelodysplastic syndromes, CMML chronic myelomonocytic leukemia, MPN myeloproliferative neoplasms, AITL angioimmunoblastic T-cell leymphoma, PTCL-NOS peripheral T-cell lymphoma, not otherwise specified, DLBCL diffuse large B-cell lymphoma, MCL mantle cell lymphoma. The rightmost column for AML indicates TET2 mutation frequencies in secondary AML (single asterisk). The diagnosis of PTCL-NOS is based on the WHO classification before the revision proposed in 2016 (double asterisk)
Following this initial discovery, recurrent TET2 mutations were also found in 2011 in subtypes of mature T/NK-cell neoplasms, particularly in T cell lymphoma with feature of follicular helper T (TFH) cells, such as angioimmunoblastic T-cell lymphoma (AITL) [16]. Subsequently, the frequency was revealed to be higher in these subtypes, particularly in AITL (>80%), than in myeloid malignancies [17–19]. Furthermore, TET2 was shown to be recurrently mutated in mature B-cell lymphomas such as diffuse large B-cell lymphoma [20] and mantle cell lymphoma [21], although at low frequencies (Fig. 2). All of these mutations are similarly distributed in the coding region and thus considered to be loss-of-function mutations.
TET2 mutations in healthy individuals
The concept of clonal hematopoiesis was established by three recent studies using cohorts of dementia [22], hypertension and coronary diseases [23], and nonhematologic malignancies [24]. In these studies, it was demonstrated that blood cell clones with leukemia-driver mutations are observed in those who do not have hematologic malignancies or clonal blood diseases. These clonal blood cells were detected at frequencies of 5% in individuals in their 60s and 10–15% of those in their 70s, with a minor diversity among the cohorts. The incidence of clonal hematopoiesis is thus clearly age-dependent. The presence of clonal blood cells was shown to be a risk factor for the development of hematologic malignancies.
After DNA methyltransferase 3A (DNMT3A), the TET2 gene is the second or the third most frequently mutated gene in these clonal hematopoiesis. This indicates that TET2 mutation per se does not induce hematologic malignancies, but rather does so in collaboration with other driver mutations.
Lessons from mice with modified or deleted Tet2 gene
Mice with knocked-down or conditionally knocked-out Tet2 have provided some additional insights into the effects of impaired TET2 function on hematopoietic cells [16, 25–30]. In general, these mice do not show evident abnormalities before reaching advanced age. However, hematopoietic stem and early progenitor cell (HSPC) fractions are increased in the bone marrow several months after the deletion of Tet2 by the induction of Cre. Moreover, when bone marrow cells from these mice are transplanted to syngenic mice, a higher chimerism is achieved than by wild-type mice-derived bone marrow cells.
Along with aging, conditional knock-out mice show a dominant increase in myeloid cells. Development of CMML-like disease has also been described in several reports [16, 25–30] These findings are thought to model, in part, myeloid malignancies in patients.
In addition, several abnormalities in T-lineage cells have also been described. In a report analyzing Tet2 knock-down mice, an increase in TFH was observed in the spleen at 20 weeks after birth, and lymphoma-like tumors developed at 60 weeks of age or older, demonstrating swollen lymph nodes, enlarged spleen, and infiltration of tumor cells into liver and lungs. The growing/infiltrating cells showed phenotypes of TFH cells [30].
Taken together, these findings indicate that it is likely that impaired 5mC-to-5hmC conversion confers clonal dominance to HSPC and exerts differentiation pressure toward the myeloid lineage. In addition, T-lineage cells may experience differentiation or growth pressure toward the TFH cell lineage.
The effects of the impaired 5mC to 5hmC conversion on B-cell lineage remain equivocal. TET2 mutations are found in various lineages of circulating blood cells, including B cells, identical to those in the tumor tissue-derived DNA from the corresponding patients with T-cell lymphoma [16, 31, 32]. This may indicate a neutral effect on B cells. Nevertheless, the development of B-cell acute lymphoblastic leukemia was recently reported in mice with double deletion of Tet1 and Tet2 [33].
TET2 mutations and pre-leukemia/pre-lymphoma
In patients with AML at remission, DNMT3A mutations, but not co-existing mutations in leukemia cells at the presentation, were identified in purified fully functional hematopoietic stem cell (HSC), progenitor cell, and mature cell fractions in peripheral blood [34]. Similarly, TET2 mutations were identified in flow cytometry (FCM)-separated residual HSCs in patients with AML; the unseparated fractions contained massive numbers of leukemia cells carrying TET2 and other mutations [35]. This suggests the FCM-separated fraction represented pre-leukemia cells.
Similarly, pre-lymphoma cells can be identified in the bone marrow and peripheral blood of patients with AITL and T-cell lymphoma with TFH cell features [16]. In these patients, unseparated bone marrow cells, colony-forming cells, and FCM-sorted monocytes as well as various lymphocytes from blood, have been shown to carry TET2 mutations identical to those discovered in the corresponding tumor tissues. DNMT3A mutations were also similarly identified, although less frequently than TET2 mutations.
All this indicates that TET2, as well as DNMT3A, mutations are acquired at the HSPC level, although these cells retain the capacity to differentiate into variety of mature blood cells that are apparently normal. Cells carrying such mutations, however, must be predisposed to subsequently acquire mutations at a variety of differentiation stages in distinct cell lineages. These cells may also act as multi-potential pre-leukemia/pre-lymphoma cells, with a tendency toward AML in DNMT3A mutations and MDS and AITL in TET2 mutations (Fig. 3).
Concept of development of different types of blood cancers from common pre-leukemia/pre-lymphoma cells. A hematopoietic stem cell (HSC) acquires a TET2 mutation. The TET2-mutated HSCs have only mild growth advantage and keep a relative proportion in the HSC compartment. Blood cells in multiple lineages are the mixture of those carrying TET2 mutations and those without. After cells in different lineages acquire an additional distinct genetic hit, these cells eventually develops into different blood cancers
Further lessons from mice with combinatorial Tet2 deletion and other blood cancer driver gene abnormalities: modeling clinical cancers. As outlined above, it is likely that TET2 mutations act as a strong initiator of hematologic malignancies, beyond the myeloid and lymphoid lineages. Nevertheless, gene mutations that are specific to individual blood cancers, such as EZH2 and ASXL1 mutations in MDS, FLT3-ITD mutations in AML, and JAK2-V1617F mutations in MPN, have been identified. These abnormalities have been modeled by gene knock-out, transgene expression, knock-in methods, and the resultant mice crossed with Tet2 knock-out mice. The offspring of such crosses have been shown to develop blood cancers recapitulating MDS [36, 37], AML [38], and MPN [39, 40]. These mouse models suggest that specific gene mutations induce development of full-blown individual blood cancers from pre-leukemia cells having TET2 mutations. Further, it has been suggested that these specific gene abnormalities determine the specificity of blood cancers.
Epigenetic abnormalities caused by TET2 mutations
Thus far, methylation profiles in cancers have mainly been characterized in CpG islands or in promoter regions. Genome-wide methylation analysis with blood cancer samples having TET2 mutations is no exception. Our understanding of the effect of TET2 mutations thus remains incomplete. It has been shown, however, that in embryonic stem cells, Tet2 single [41] and Tet1/2/3 triple [42] knock-out results in hypermethylation at selected enhancer regions.
The effect of Tet2 on enhancer methylation in the hematopoietic compartment has also been demonstrated in a small number of studies. When the AML1-ETO fusion protein was expressed in Tet2 conditionally knocked-out granulocyte–macrophage progenitors, progressive hypermethylation of active enhancer elements was observed during the transformation of the cells [43]. This specific enhancer hypermethylation phenotype has also been found in human AML cells carrying TET2 mutations.
In a TFH-like lymphoma model using Tet2 knock-down mice, an intronic enhancer in the Bcl6 gene was hypermethylated (Fig. 4), and Bcl6 expression was upregulated. In this model, Bcl6 protein was highly expressed in the tumor cells [30]. BCL6 is an essential transcription factor in TFH cell development, and its expression was thus presumed to contribute to the outgrowth of TFH cells.
Differential methylation in an intronic silencer in Bcl6 gene. KD/KD, Tet2-knock-down mice. Bisulfite sequencing at the CpG-rich region at the first intron of Bcl6 gene in splenic CD4(+) cells from wild-type and KD/KD mice. Adopted from Ref. [30] with a modification
A recent analysis of mice with double deletion of Tet2 and Dnmt3a genes proposes a more precise and complex model of the regulation of methylation, in which the two DNA methylation regulators show competitive as well as cooperative and independent actions on the methylome [44]. The presence of 5-hmC in the gene body served by Tet2 is important to the maintenance of HSC-specific gene expression, to which the contribution of Dnmt3a is minor. Repression/primed state of various lineage-specific genes is cooperatively repressed by the co-presence of 5mC and 5hmC at the transcription start sites (Fig. 5). Double deletion induces expression of these lineage-specific genes such as Krueppel-like factor 1 (Klf1), a transcription factor, and erythropoietin receptor (Epor), a signaling molecule. This may help HSC to grow by enhanced mitotic signaling, while it does not induce terminal differentiation.
Primed state in hematopoietic stem cells. The presence of both 5-mC and 5-hmC is necessary to maintain the primed state. If both DNMT3A and TET2 are impaired, lineage-specific transcription factors are aberrantly expressed in HSCs. Based on Ref. [44]
Concluding remarks
The scheme of pre-leukemia/pre-lymphoma due to defective function by mutations in TET2 and DNMT3A has been getting clarified. Although the landscape of DNA methylome and hydroxymethylome remains hazy, the fog is gradually lifting. The current technique used in methylome analysis, i.e., bisulfite sequencing, may represent a bottleneck, as this technology cannot distinguish 5-mC from 5-hmC. Comprehensive elucidation of the function of TET2 and other DNA methylation regulators will remain an important question for future research.
References
Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–66.
Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–5.
Franchini DM, Schmitz KM, Petersen-Mahrt SK. 5-Methylcytosine DNA demethylation: more than losing a methyl group. Annu Rev Genet. 2012;46:419–41.
Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14(6):341–56.
Ko M, Huang Y, Jankowska AM, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468(7325):839–43.
He YF, Li BZ, Li Z, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–7.
Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12(9):599–612.
Sakata-Yanagimoto M, Enami T, Yokoyama Y, Chiba S. Disease-specific mutations in mature lymphoid neoplasms: recent advances. Cancer Sci. 2014;105(6):623–9.
Delhommeau F, Dupont S, Della Valle V, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360(22):2289–301.
Langemeijer SM, Kuiper RP, Berends M, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41(7):838–42.
Jankowska AM, Szpurka H, Tiu RV, et al. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood. 2009;113(25):6403–10.
Tefferi A, Pardanani A, Lim KH, et al. TET2 mutations and their clinical correlates in polycythemia vera, essential thrombocythemia and myelofibrosis. Leukemia. 2009;23(5):905–11.
Haferlach T, Nagata Y, Grossmann V, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241–7.
Fernandez-Mercado M, Yip BH, Pellagatti A, et al. Mutation patterns of 16 genes in primary and secondary acute myeloid leukemia (AML) with normal cytogenetics. PLoS One. 2012;7(8):e42334.
Hu L, Li Z, Cheng J, et al. Crystal structure of TET2-DNA complex: insight into TET-mediated 5mC oxidation. Cell. 2013;155(7):1545–55.
Quivoron C, Couronné L, Della Valle V, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20(1):25–38.
Sakata-Yanagimoto M, Enami T, Yoshida K, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46(2):171–5.
Lemonnier F, Couronné L, Parrens M, et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood. 2012;120(7):1466–9.
Wang C, McKeithan TW, Gong Q, et al. IDH2R172 mutations define a unique subgroup of patients with angioimmunoblastic T-cell lymphoma. Blood. 2015;126(15):1741–52.
Asmar F, Punj V, Christensen J, et al. Genome-wide profiling identifies a DNA methylation signature that associates with TET2 mutations in diffuse large B-cell lymphoma. Haematologica. 2013;98(12):1912–20.
Meissner B, Kridel R, Lim RS, et al. The E3 ubiquitin ligase UBR5 is recurrently mutated in mantle cell lymphoma. Blood. 2013;121(16):3161–4.
Genovese G, Kähler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477–87.
Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–98.
Xie M, Lu C, Wang J, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472–8.
Moran-Crusio K, Reavie L, Shih A, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20(1):11–24.
Ko M, Bandukwala HS, An J, et al. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci USA. 2011;108(35):14566–71.
Li Z, Cai X, Cai CL, et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118(17):4509–18.
Shide K, Kameda T, Shimoda H, et al. TET2 is essential for survival and hematopoietic stem cell homeostasis. Leukemia. 2012;26(10):2216–23.
Kunimoto H, Fukuchi Y, Sakurai M, et al. Tet2 disruption leads to enhanced self-renewal and altered differentiation of fetal liver hematopoietic stem cells. Sci Rep. 2012;2:273.
Muto H, Sakata-Yanagimoto M, Nagae G, et al. Reduced TET2 function leads to T-cell lymphoma with follicular helper T-cell-like features in mice. Blood Cancer J. 2014;4:e264.
Couronné L, Bastard C, Bernard OA. TET2 and DNMT3A mutations in human T-cell lymphoma. N Engl J Med. 2012;366(1):95–6.
Nguyen TB, Sakata-Yanagimoto M, Nakamoto-Matsubara R, et al. Double somatic mosaic mutations in TET2 and DNMT3A–origin of peripheral T cell lymphoma in a case. Ann Hematol. 2015;94(7):1221–3.
Zhao Z, Chen L, Dawlaty MM, et al. Combined loss of Tet1 and Tet2 promotes B cell, but not myeloid malignancies in mice. Cell Rep. 2015;13(8):1692–704.
Shlush LI, Zandi S, Mitchell A, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506(7488):328–33.
Jan M, Snyder TM, Corces-Zimmerman MR, et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med. 2012;4(149):149ra118.
Muto T, Sashida G, Oshima M, et al. Concurrent loss of Ezh2 and Tet2 cooperates in the pathogenesis of myelodysplastic disorders. J Exp Med. 2013;210(12):2627–39.
Abdel-Wahab O, Gao J, Adli M, et al. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J Exp Med. 2013;210(12):2641–59.
Shih AH, Jiang Y, Meydan C, et al. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer Cell. 2015;27(4):502–15.
Chen E, Schneider RK, Breyfogle LJ, et al. Distinct effects of concomitant Jak2V617F expression and Tet2 loss in mice promote disease progression in myeloproliferative neoplasms. Blood. 2015;125(2):327–35.
Kameda T, Shide K, Yamaji T, et al. Loss of TET2 has dual roles in murine myeloproliferative neoplasms: disease sustainer and disease accelerator. Blood. 2015;125(2):304–15.
Hon GC, Song CX, Du T, et al. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol Cell. 2014;56(2):286–97.
Bogdanović O, Smits AH, de la Calle Mustienes E, et al. Active DNA demethylation at enhancers during the vertebrate phylotypic period. Nat Genet. 2016;48(4):417–26.
Rasmussen KD, Jia G, Johansen JV, et al. Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev. 2015;29(9):910–22.
Zhang X, Su J, Jeong M, et al. DNMT3A and TET2 compete and cooperate to repress lineage-specific transcription factors in hematopoietic stem cells. Nat Genet. 2016;48(9):1014–23.
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Chiba, S. Dysregulation of TET2 in hematologic malignancies. Int J Hematol 105, 17–22 (2017). https://doi.org/10.1007/s12185-016-2122-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12185-016-2122-z