Abstract
Posttranscriptional gene regulation by small RNAs (15–40-nucleotide noncoding RNAs) is now established as an important branch of the gene regulatory system. It has recently been revealed that noncoding RNAs can be categorized into different types and that they work through novel mechanisms. In addition, it has been shown that noncoding RNAs mediate intercellular communication and, importantly, that cross talk between coding and noncoding RNAs occurs. In this review, we discuss the recent findings concerning small RNAs. It was originally proposed that microRNAs (miRNAs) work to “fine tune” the determination of cell fate. However, critical functions beyond fine tuning have been revealed. In addition to miRNAs, next-generation sequencing has revealed the existence of various species of non-canonical small RNAs: mirtrons, piRNAs, 21U-RNA, endo-siRNAs, snoRNAs, usRNAs, and Y-RNA-derived small RNAs. Some of these species are involved in response to viral infection. Finally, we highlight the intracellular functions of small RNAs, which involve the exosomes.
Similar content being viewed by others
microRNAs as fine tuners of hematopoiesis
It is well known that transcription factors are one of the critical determinants of cell fate (Table 1). In hematopoiesis, a variety of transcription factors [1–4] and epigenetic regulators [5–9] have been identified as master regulators/genes for each hematopoietic cell lineage. Recently, many researchers have focused on microRNAs (miRNAs) as modulators—so-called fine tuners—of the expression of various genes, including transcription factors, during cell differentiation. miRNAs are a class of noncoding small RNAs that posttranscriptionally regulate gene expression. Single miRNA species regulate the expression of various target mRNAs by suppressing translation or stabilizing the mRNA.
miR-150 is one of the most documented miRNAs involved in the development of megakaryocytes and erythrocytes. The concentration of the transcription factor Myb in megakaryocyte–erythrocyte precursor cells critically affects the miRNA’s fate [10]. Abundant Myb promotes growth toward the erythrocyte lineage, whereas lower expression levels lead to megakaryocyte development [10]. Myb is an evolutionally conserved target of miR-150 [11, 12]. By regulating the amount of Myb in precursor cells, miR-150 directs them toward the megakaryocyte lineage [11]. miR-150 has also been reported to regulate the development of B cells; expansions of B-1 cells were observed in miR-150-deficient mice [12]. Conversely, miR-150 overexpression impaired transition of pro-B cells to the pre-B stage, similarly to that observed in the heterozygous Myb mutated mouse [12]. These observations point toward an important role of miR-150 as a regulator of cell differentiation via the maintenance of Myb expression at its optimal level.
Other miRNAs have been widely observed to perform multiple tasks in various cell lineages. miR-181a has been reported to promote the expression of B and natural killer (NK) cell genes [13, 14]. In T-lymphopoiesis, miR-181a affects selection of the T cell repertoire by modulating the intensity of signaling via T cell receptors [15, 16]. More recently, miR-181a has been reported to have a function in NKT cells [17, 18].
miR-146a is a multifunctional miRNA involved in hematopoiesis. The expression of miR-146a is high in hematopoietic stem/precursor cells and myeloid precursor cells [19]. Enforced expression of miR-146a in hematopoietic stem cells (HSCs) facilitates myelopoiesis, which is accompanied by a reduction in lymphoid and erythroid differentiation [19]. At a later stage, miR-146a promotes the development of peritoneal macrophages [20]. Megakaryocytopoiesis is also under the control of miR-146a. Enforced expression of miR-146a suppresses the ability of hematopoietic precursor cells (HPCs) to give rise to mature megakaryocytes, and the antagonism of miR-146a results in the upregulation of megakaryocyte-related genes [20]. Moreover, miR-146a was first reported as a key factor modulating the innate immune response. Fas is a direct target of miR-146a. Therefore, Fas suppression by miR-146a represents a major etiological factor for pathological manifestation of human autoimmune lymphoproliferative syndrome (ALPS). This downregulation pattern suggests a binary off-switch to decrease the Fas expression in GC B cells. Moreover, miR-146a functions as a robust “brake” during the inflammation and adoptive immune responses. miR-146 is abundantly present in various different lymphocytes subsets such as Tfh, Treg and germinal center B cells [21].
Co-operative regulation of the differentiation of single cell lineages by multiple miRNAs is usually observed. In addition to miR-181a and miR-150, the miR-17–92 cluster [22], miR-34a [23] and miR-125 [24] have also been reported in B-lymphopoiesis. miR-181a and the miR-17–92 cluster were reported to be positive regulators of B-lymphopoiesis, whereas miR-34a, miR-150, and miR-125 negatively regulate the generation of B cells. miR-125b binds to the B lymphocyte-induced maturation protein-1 (BLIMP-1) and IFN regulatory protein-4 (IRF-4) transcription factors and regulates plasma cell differentiation [25] and overexpression of miR-125b protected pre-BI and leukemic B cell lines from apoptosis [26]. The miR-17–92 cluster consists of six miRNAs and plays a role in the transition of pro-B to pre-B cells. Downregulation of a pro-apoptotic gene, Bim, by miR-17–92 has been suggested to regulate apoptosis of pro-B cells at this checkpoint. At the same transition point, miR-34a modulates Foxp1 expression. Foxp1 is a transcription factor that is highly expressed in diffuse large B lymphoma and is essential for B-lymphopoiesis [27]. Lack of Foxp1 leads to a defect in B cell differentiation, and overexpression of miR-34a in bone-marrow cells mimics Foxp1 deficiency.
MiRNAs are involved in the control of HSC quiescence [28, 29] and activation. MiR-126, which is highly expressed in HSCs and early hematopoietic progenitors, plays a pivotal role in restraining cell-cycle progression of HSCs in vitro and in vivo. miR-126 knockdown, using lentiviral sponges, increased HSC proliferation without inducing exhaustion, resulting in expansion of mouse and human long-term repopulating HSCs. Conversely, enforced miR-126 expression impairs cell-cycle entry, leading to progressively reduced hematopoietic contribution. In HSCs/early progenitors, miR-126 regulates multiple targets within the PI3 K/AKT/GSK3β pathway, attenuating signal transduction in response to extrinsic signals. These data establish that miR-126 sets are a threshold for HSC activation and therefore governs HSC pool size, demonstrating the importance of miRNAs in the control of HSC function [30].
microRNAs as more than fine tuners of hematopoiesis
At the end of this section, we propose that miRNAs might act as on/off switches rather than fine tuners in some cases. E12/E47 (E2A), early B cell factor 1 (EBF1), and PAX5 are critical transcription factors in B-lymphopoiesis. EBF1 is indispensable for development of the B cell lineage (Fig. 1) [27, 31]. Deletion of either of EBF1, E2A, or PAX5 impairs B-lymphopoiesis at the stage of commitment. The suppression of B-lymphopoiesis caused by the absence of E2A can be countered by the expression of EBF, but not by that of PAX5. The impairment caused by the absence of PAX5 can be partially overcome by EBF, but not by that of E2A, while the impairment caused by the deficiency of EBF can be countered by neither E2A nor PAX5 expression, indicating that EBF is the most important transcription factor required for development of the B cell lineage.
Recently, we reported that miR-126 drives MLL-AF4 acute lymphocytic leukemia (ALL) cells, exhibiting both myeloid and B cell surface markers, toward B cells without any upregulation of EBF1, E2A, or PAX5 [32]. MiR-126 also increases B cell differentiation of normal HPCs, and this function was prominently observed in the uncommitted HPCs lineage (ckit+ Sca1+ cells), indicating that miR-126 is involved in cell fate determination, but not in B cell commitment. These results suggest that miRNAs can induce B cell commitment independently of critical transcription factors, such as EBF1, E2A, and PAX5. Using miR-126, we partially recovered the potential for differentiation along the B cell lineage in HPCs lacking EBF1 (Fig. 2). These findings suggest that miRNAs might be capable of inducing cell differentiation via a transcription factor-independent pathway, which means that miRNAs function as more than fine tuners in cell fate determination, at least in B cell lineage commitment. Moreover, the observed function of miR-126 as an inducer of differentiation suggests that miR-126 might be a promising therapeutic target. In acute leukemia, inhibition of differentiation caused by deregulation of transcription factors is assumed to be a critical factor for the development of leukemia [33–36]. MiR-126 might restore function and be a potential agent for differentiation induction therapy for ALL. Thus, further studies are needed to evaluate its potential as a differentiation inducer.
a miR-126 constructed on retroviral vector carrying rat CD2 was transduced into Ebf1 −/− HPCs. Cells were cultured for 4 weeks, and the expressions of B220 and CD19 were evaluated by flow-cytometry on every other week. b Expressions of E2A, Pax5, CD79a, CD79b, Rag1, Rag2, Vpreb1 and Igll1 in empty (Ctrl.) or miR-126 vector-transduced cells were determined by Agilent gene expression arrays
Non-canonical small RNAs and viral infection
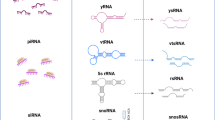
Recent innovations in sequencing technology have brought the discovery of small noncoding RNAs, including miRNAs, mirtrons, piRNAs, 21U-RNA, endo-siRNAs, snoRNAs, usRNAs [37] and Y-RNA-derived small RNAs [38], which appear to be involved in many critical biological processes (Fig. 3).
Cell-to-cell communication with exosomes: a when some kinds of tumor cells are activated by stimulation and so on, exosomes which contain microRNAs are secreted from activated tumor cells. The exosomes are taken by non-tumor cells like macrophages. b MicroRNAs transferred through exosomes can induce inflammation, cell-proliferation and so on in donor cells. In some cases, the secretion of exosomes is dysregulated and chronic inflammation is induced in non-tumor cells. The abnormal non-tumor cells accelerate proliferation of tumor cells
Mirtrons are miRNAs that regulate RNA via a multistep process involving intron splicing and debranching, exosome-mediated trimming of the 3-tail, and dicing. Mirtrons can be further categorized [39, 40] into typical mirtrons, which are derived from the whole regions of short introns, and tailed mirtrons, which are generated from end regions of longer introns [41, 42]. Mammalian studies have identified three well-conserved mirtrons. The most prominent example is miR-877, a typical mirtron whose miRNA and miRNA sequences reside near the exon/intron boundaries. In the hematopoietic system, the mirtron miR-1,233 was detected in childhood ALL [38].
In contrast to miRNAs, endo-siRNAs are derived from long double-stranded RNAs (dsRNAs) in the form of annealed natural antisense transcripts (NATs) or long hairpin RNAs (hp-RNAs). Processing of long dsRNAs is directly performed by Dicer to consecutively produce multiple siRNAs. Diverse sources of dsRNA have been identified in animals and plants. Multiple species of 21–22-nucleotide endo-siRNAs can be generated via the sequential cleavage activity of Dicer along a long hairpin. Although miRNAs are widely expressed in various cells and tissues, currently endo-siRNAs have only been reported in mouse stem cells and oocytes [41, 43–45].
PIWI-interacting RNAs (piRNAs) are derived from transcripts of piRNA loci, processed by PIWI family proteins, which suppress target gene transcription via methylation of retrotransposons [46, 47]. piRNAs that modulate spermatogenesis in mammals, piRNA-like repeat-associated siRNAs that regulate chromatin structure of retrotransposons in insect germ lines, transcription start-site-associated RNAs (TSSa-RNAs), and the functionally uncharacterized piRNA-like 21U-RNAs harboring 5-uredines were discovered in worms [48–51]. Another type of non-canonical small RNA originates from small nucleolar RNAs (snoRNAs) through their internal hairpin-shaped folding structures [52]. Processing of snoRNA-derived hairpins gives rise to Dicer-dependent but Drosha/Dgcr8-independent miRNAs. Besides snoRNAs, transfer RNA (tRNA) transcripts might also fold into alternative hairpin structures, on which Dicer exerts cleavage activities without the processing of Drosha/Dgcr8 [41]. Other non-canonical small RNAs arise from endogenous short hairpin RNAs (shRNAs), from which transcripts originating from unannotated, intergenic regions can serve as Dicer substrates upon transcription. In the processing of miR-451, which is highly expressed in erythrocytes, miRNA biogenesis depends on Drosha, but bypasses Dicer processing where Argonaute (AGO)-containing RNA-induced silencing complex (RISC) substitutes Dicer for the processing of pre-miRNAs [53–55].
Kaposi sarcoma-associated herpes virus K12-1 miRNA (23 bases) associates with a distinct, unusually small RNA (usRNA; 17 bases) that can effectively downregulate a K12-1 miRNA target, human RAD21, suggesting that stable degradation-like products might also contribute to gene regulation [56]. usRNAs are shorter than canonical miRNAs. A subset of miRNAs produces usRNAs specifically from their 5-ends, which are also associated with Ago proteins and manifest a stronger preference for Ago2 than for Ago1. miRNA-derived usRNAs are found in JC virus (miR-J1) and Huh7 cells (miR-122) [57, 58]. Cumulatively, miRNA-derived usRNAs might also contribute to gene regulation. In addition to miRNA-derived usRNAs, 15–18-base non-miRNA-derived usRNAs have been found, which harbor motifs that are both over-represented and highly preferred at precise locations on their 3 ends [56]. Intriguingly, all three position-specific motifs that are overrepresented in usRNAs and the motif detected in Ago-associated 15–18-base RNAs are C-riched. The absence of such motifs in the large class of TSSa-RNAs and piRNAs suggests that these motifs do not simply increase the stability (e.g., exonuclease resistance) of RNA transcripts. Independent detection of similar motifs in Ago-associated non-miRNA-derived usRNAs further highlights the importance of such motifs.
Y-RNA-derived miRNA was detected in a virus-infected cell. The expression of Y-RNA-derived has-miR-1975 and has-miR-1979 was significantly upregulated in enterovirus 71 (EV71)-infected Hep2 (human epidermal carcinoma) cells, which cause hand, foot, and mouth disease (HFMD) [59]. These non-canonical small RNAs were also upregulated in adenovirus type 3 (AD3)-infected Hep2 cells. These studies suggest that the two Y-RNA-derived small RNAs are involved in virus infection [60].
miRNAs and siRNAs are incorporated into the multiprotein RISC, exerting posttranscriptional repression of target mRNAs. In lower organisms, the siRNA and miRNA pathways diverge, in part due to sorting mechanisms that direct distinct small RNA duplexes into specific Ago-RISCs [61, 62]. However, such sorting mechanisms appear to be lost in mammals. Ago1–4 appear to be indistinguishable for miRNAs, whereas Ago1 and Ago2 are preferred by siRNAs. By contrast, influenza-A-virus-infected Ago1 and Ago3 double-knockout mice exhibited increased mortality, consistent with more severe alveolitis and pneumonitis, indicating that optimal resistance to influenza requires Ago1 and/or Ago3 [63]. Enhanced mortality of double-knockout mice was associated with neither increased viral replication nor differential pulmonary recruitment or function of innate and adaptive immune cells; it remains to be shown whether its function includes the RNA interference, which targets the virus-coding RNAs. Experiments showed that Ago1–4 appear to be indistinguishable for miRNAs, but distinct for other small RNAs.
Recently, we found small RNAs preferentially associated with Ago1 in Epstein–Barr virus (EBV)-infected cells. Interestingly, the unique reads were aligned on transfer RNAs, mitochondrial transfer RNAs, 5S ribosomal RNA, snoRNAs, and snRNAs. We named this family of small RNAs preferentially associated with Ago1, Agotaxis small RNAs (Agotaxis sRNAs; ASRs). Quantitative PCR analysis revealed that the Agotaxis sRNAs were expressed not only in EBV-positive cell lines, but also in EBV-negative cells, suggesting that the Agotaxis sRNAs are expressed regardless of EBV infection. However, the Agotaxis sRNAs were dramatically upregulated in EBV lytic infection. Moreover, when we knocked down Ago1 at this phase, Agotaxis sRNAs were reduced. In addition, apoptosis, which is the terminal phase of lytic infection, was upregulated. These results indicate that upregulation of Agotaxis sRNAs is dependent on AGO1 and suppresses apoptosis. Agotaxis sRNAs might have crucial functions through the posttranscriptional regulation of target genes to prevent the lytic infection from “overactivation”.
Secreted miRNAs are novel intracellular communicators
Multicellular organisms require cell-to-cell communication for appropriate development and to maintain homeostasis, among other things. Classic cell-to-cell communication can be mediated by cell junctions, adhesion contacts, or soluble messengers [64]. The soluble factors include hormones, cytokines, and chemokines that bind to specialized receptors expressed on the surface of target cells. In addition to the well-studied signaling pathways, many other mechanisms of intercellular communication have now been proposed, including secreted miRNAs [65].
There are two pathways for active secretion of miRNAs—exosome-dependent and RNA-binding-protein-associated pathways [66]. Exosomes are small secreted vesicles with a diameter of 30–100 nm and contain a variable spectrum of parental cell molecules, including RNAs, proteins, and lipids [67]. Cell recognition molecules on exosome surfaces or specific receptors on recipient cell membranes could allow specificity in exosome uptake [68]. Interestingly, miRNA packaging occurs non-randomly, and specific miRNA populations are preferentially sorted into exosomes. Valadi et al. and Skog et al. [68, 69] showed that certain miRNAs were present dominantly or at higher levels in exosomes than in the donor cells. Furthermore, Zomer et al. [70] discovered that purified exosomes contain functional miRNAs and small RNAs, but detected little mRNA. They speculate that exosomes are specialized in carrying small RNA including the 22–25-nucleotide regulatory miRNAs. In addition to miRNA secretion, many researchers have also demonstrated uptake of secreted miRNAs by recipient cells and their physiological effects [66]. Therefore, secreted miRNAs represent a newly discovered mechanism by which donor cells can influence the gene expression of recipient cells.
Recent studies revealed that horizontal transfer of exosomal miR-210 from metastatic cancer cells could modulate the microenvironmental endothelial cells for the benefit of the cancer cells, which contributed to cancer metastasis. Preventing the expression of nSMase2, which regulates exosomal miRNA secretion, in metastatic cancer cells abrogates the metastatic ability of cancer cells to target lung tissues, whereas reconstitution via the administration of exosomes isolated from metastatic cancer cells rescued this phenomenon. In this context, the number of endothelial cells in inoculated tumors was proportional to the expression level of nSMase2 in cancer cells. In fact, exosomes derived from a metastatic cancer cell line enhanced the capillary formation and migration of endothelial cells in vitro. Interestingly, the expression profiles of exosomal miRNAs obtained from metastatic cancer cells demonstrated that a set of angiogenic miRNAs were highly concentrated in these exosomes. One of them, miR-210, enhanced angiogenesis through the suppression of specific target genes. These results revealed that cancer cells provide nSMase2-regulated exosomal miRNAs to endothelial cells to promote their metastatic initiation efficiency.
EBV is the first known virus to encode miRNAs, and 44 mature EBV-miRNAs have been reported [71]. EBV generates a variety of miRNAs by processing the BHRF1 and BamHI A rightward (BART) transcripts [72]. EBV-miRNAs can regulate viral and cellular targets, and they generally target host transcripts during latent infection [73]. For example, EBV-miR-BART15-3p targets baculovirus inhibitor of apoptosis repeat-containing ubiquitin-conjugating enzyme (BRUCE) [72]. Choi et al. suggested that EBV-miR-BART15-3p can induce apoptosis partially by inhibiting the translation of the apoptosis inhibitor BRUCE. Moreover, some EBV-miRNAs share seed sequence homology with endogenous miRNAs. For example, EBV-miR-BART5 and EBV-miR-BART1-3p share similarity with endogenous miR-18 and miR-29a/b, respectively [74, 75]. BART miRNAs are expressed in all cells latently infected with EBV; however, the functions of most BART miRNAs remain unknown.
EBV-miRNAs are abundantly expressed in EBV-associated tumors, such as diffuse large B cell lymphoma (DLBCL) and nasopharyngeal carcinoma (NPC) [76]. Importantly, EBV-miRNAs secreted by EBV-infected cells are transferred to, and act in, uninfected recipient cells through exosomes [77]. Pegtel et al. reported that these EBV-miRNAs repressed the confirmed target gene chemokine (C-X-C motif) ligand 11 (CXCL11), a small chemokine belonging to the CXC chemokine family, which interacted with the cell surface chemokine receptor CXCR3 and induced a chemotactic response in activated T cells [77, 78].
Two other studies also reported that exosomes released from EBV-positive NPC cell lines contain viral miRNAs [76, 79]. Moreover, Zomer et al. [70] detected EBV-miRNAs in non-infected cells in the peripheral blood of patients that include monocytes and T cells. These studies reveal that miRNAs secreted by EBV-infected cells can be transferred to uninfected recipient cells and silence immunoregulatory genes in the recipient cells.
The elucidation of this novel information transfer system will be important in understanding many biological processes including development, immune response, and tumorigenesis [66]. The miRNA composition in secreted exosomes might reflect molecular changes in donor cells and therefore may provide diagnostic information and aid in therapeutic decisions for cancer patients.
References
Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21(21):3368–76.
Friedman AD. Transcriptional regulation of myelopoiesis. Int J Hematol. 2002;75(5):466–72.
Ye M, Graf T. Early decisions in lymphoid development. Curr Opin Immunol. 2007;19(2):123–8.
Zhu J, Emerson SG. Hematopoietic cytokines, transcription factors and lineage commitment. Oncogene. 2002;21(21):3295–313.
Sashida G, Iwama A. Epigenetic regulation of hematopoiesis. Int J Hematol. 2012;96(4):405–12.
Chung YR, Schatoff E, Abdel-Wahab O. Epigenetic alterations in hematopoietic malignancies. Int J Hematol. 2012;96(4):413–27.
Zhang Y, Chen A, Yan XM, Huang G. Disordered epigenetic regulation in MLL-related leukemia. Int J Hematol. 2012;96(4):428–37.
Yamazaki J, Issa JP. Epigenetic aspects of MDS and its molecular targeted therapy. Int J Hematol. 2013;97(2):175–82.
Milosevic JD, Kralovics R. Genetic and epigenetic alterations of myeloproliferative disorders. Int J Hematol. 2013;97(2):183–97.
Emambokus N, Vegiopoulos A, Harman B, Jenkinson E, Anderson G, Frampton J. Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. EMBO J. 2003;22:4478–88. doi:10.1093/emboj/cdg434.
Lu J, Guo S, Ebert BL, Zhang H, Peng X, Bosco J, Pretz J, Schlanger R, Wang JY, Mak RH, Dombkowski DM, Preffer FI, Scadden DT, Golub TR. MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev Cell. 2008;14:843–53. doi:10.1016/j.devcel.2008.03.012.
Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–59. doi:10.1016/j.cell.2007.07.021.
Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–6. doi:10.1126/science.1091903.
Cichocki F, Felices M, McCullar V, Presnell SR, Al-Attar A, Lutz CT, Miller JS. Cutting edge: microRNA-181 promotes human NK cell development by regulating Notch signaling. J Immunol. 2011;187:6171–5. doi:10.4049/jimmunol.1100835.
Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, Klein LO, Davis MM, Chen CZ. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–61. doi:10.1016/j.cell.2007.03.008).
Ebert PJ, Jiang S, Xie J, Li QJ, Davis MM. An endogenous positively selecting peptide enhances mature T cell responses and becomes an autoantigen in the absence of microRNA miR-181a. Nat Immunol. 2009;10:1162–9. doi:10.1038/ni.1797).
Ziętara N, Łyszkiewicz M, Witzlau K, Naumann R, Hurwitz R, Langemeier J, Bohne J, Sandrock I, Ballmaier M, Weiss S, Prinz I, Krueger A. Critical role for miR-181a/b-1 in agonist selection of invariant natural killer T cells. Proc Natl Acad Sci USA. 2013;110:7407–12. doi:10.1073/pnas.1221984110).
Henao-Mejia J, Williams A, Goff LA, Staron M, Licona-Limón P, Kaech SM, Nakayama M, Rinn JL, Flavell RA. The microRNA miR-181 is a critical cellular metabolic rheostat essential for NKT cell ontogenesis and lymphocyte development and homeostasis. Immunity. 2013;38:984–97. doi:10.1016/j.immuni.2013.02.021.
Starczynowski DT, Kuchenbauer F, Wegrzyn J, Rouhi A, Petriv O, Hansen CL, Humphries RK, Karsan A. MicroRNA-146a disrupts hematopoietic differentiation and survival. Exp Hematol. 2011;39:167–78. doi:10.1016/j.exphem.2010.09.011.
Labbaye C, Spinello I, Quaranta MT, Pelosi E, Pasquini L, Petrucci E, Biffoni M, Nuzzolo ER, Billi M, Foà R, Brunetti E, Grignani F, Testa U, Peschle C. A three-step pathway comprising PLZF/miR-146a/CXCR4 controls megakaryopoiesis. Nat Cell Biol. 2008;10:788–801. doi:10.1038/ncb1741.
Guo Q, Zhang J, Li J, et al. Forced miR-146a expression causes autoimmune lymphoproliferative syndrome in mice via downregulation of Fas in germinal center B cells. Blood. 2013;121(24):4875–83.
Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–86. doi:10.1016/j.cell.2008.02.019).
Rao DS, O’Connell RM, Chaudhuri AA, Garcia-Flores Y, Geiger TL, Baltimore D. MicroRNA-34a perturbs B lymphocyte development by repressing the forkhead box transcription factor Foxp1. Immunity. 2010;33:48–59. doi:10.1016/j.immuni.2010.06.013.
Chaudhuri AA, So AY, Mehta A, Minisandram A, Sinha N, Jonsson VD, Rao DS, O’Connell RM, Baltimore D. Oncomir miR-125b regulates hematopoiesis by targeting the gene Lin28A. Proc Natl Acad Sci USA. 2012;109:4233–8. doi:10.1073/pnas.1200677109.
Gururajan M, Haga CL, Das S, et al. MicroRNA 125b inhibition of B cell differentiation in germinal centers. Int Immunol. 2010;22(7):583–92.
Puissegur MP, Eichner R, Quelen C, et al. B-cell regulator of immunoglobulin heavy-chain transcription (Bright)/ARID3a is a direct target of the oncomir microRNA-125b in progenitor B-cells. Leukemia. 2012;26(10):2224–32.
Hu H, Wang B, Borde M, Nardone J, Maika S, Allred L, Tucker PW, Rao A. Foxp1 is an essential transcriptional regulator of B cell development. Nat Immunol. 2006;7:819–26. doi:10.1038/ni1358.
Kikushige Y, Miyamoto T. TIM-3 as a novel therapeutic target for eradicating acute myelogenous leukemia stem cells. Int J Hematol. 2013;98(6):627–33.
Ishikawa F. Modeling normal and malignant human hematopoiesis in vivo through newborn NSG xenotransplantation. Int J Hematol. 2013;98(6):634–40.
Lechman ER, Gentner B, van Galen P, Giustacchini A, Saini M, Boccalatte FE, Hiramatsu H, Restuccia U, Bachi A, Voisin V, Bader GD, Dick JE, Naldini L. Attenuation of miR-126 activity expands HSC in vivo without exhaustion. Cell Stem Cell. 2012;11:799–811. doi:10.1016/j.stem.2012.09.001.
Yokota T, Sudo T, Ishibashi T, Doi Y, Ichii M, Orirani K, et al. Complementary regulation of early B-lymphoid differentiation by genetic and epigenetic mechanisms. Int J Hematol. 2013;98(4):382–9.
Okuyama K, Ikawa T, Gentner B, Hozumi K, Harnprasopwat R, Lu J, Yamashita R, Ha D, Toyoshima T, Chanda B, Kawamata T, Yokoyama K, Wang S, Ando K, Lodish HF, Tojo A, Kawamoto H, Kotani A. MicroRNA-126-mediated control of cell fate in B-cell myeloid progenitors as a potential alternative to transcriptional factors. Proc Natl Acad Sci USA. 2013;110:13410–5. doi:10.1073/pnas.1220710110).
Ito T. Stem cell maintenance and disease progression in chronic myeloid leukemia. Int J Hematol. 2013;98(6):641–7.
Chan SM, Majeti R. Role of DNMT3A, TET2, and IDH1/2 mutations in pre-leukemic stem cells in acute myeloid leukemia. Int J Hematol. 2013;98(6):648–57.
Ogawa S. Splicing factor mutations in myelodysplasia. Int J Hematol. 2012;96(4):438–42.
Naoe T, Kiyoi H. Gene mutations of acute myeloid leukemia in the genome era. Int J Hematol. 2013;97(2):165–74.
Jung CH, Hansen MA, Makunin IV, Korbie DJ, Mattick JS. Identification of novel non-coding RNAs using profiles of short sequence reads from next generation sequencing data. BMC Genomics. 2010;11:77. doi:10.1186/1471-2164-11-77).
Schotte D, Chau JC, Sylvester G, Liu G, Chen C, van der Velden VH, Broekhuis MJ, Peters TC, Pieters R, den Boer ML. Identification of new microRNA genes and aberrant microRNA profiles in childhood acute lymphoblastic leukemia. Leukemia. 2009;23:313–22. doi:10.1038/leu.2008.286).
Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi:10.1016/j.cell.2007.06.028).
Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–6. doi:10.1038/nature05983).
Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–85. doi:10.1101/gad.1705308.
Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, Blelloch R, Schroth GP, Nusbaum C, Bartel DP. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi:10.1101/gad.1884710).
Yi R, Pasolli HA, Landthaler M, Hafner M, Ojo T, Sheridan R, Sander C, O’Carroll D, Stoffel M, Tuschl T, Fuchs E. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc Natl Acad Sci USA. 2009;106:498–502. doi:10.1073/pnas.0810766105).
Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–8. doi:10.1038/nature06904.
Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, Surani MA, Sakaki Y, Sasaki H. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–43. doi:10.1038/nature06908).
Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–14. doi:10.1016/j.devcel.2007.03.001.
Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–7. doi:10.1126/science.1142612).
Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi:10.1038/nature04917.
Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–7. doi:10.1126/science.1130164).
Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, Gvozdev VA. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007;35:5430–8. doi:10.1093/nar/gkm576).
Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–207. doi:10.1016/j.cell.2006.10.040).
Ender C, Krek A, Friedländer MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–28. doi:10.1016/j.molcel.2008.10.017).
Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–9. doi:10.1038/nature09092).
Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, Wolfe SA, Giraldez AJ. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–8. doi:10.1126/science.1190809.
Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O’Carroll D, Lai EC. Conserved vertebrate miR-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci USA. 2010;107:15163–8. doi:10.1073/pnas.1006432107.
Li Z, Kim SW, Lin Y, Moore PS, Chang Y, John B. Characterization of viral and human RNAs smaller than canonical MicroRNAs. J Virol. 2009;83:12751–8. doi:10.1128/JVI.01325-09.
Seo GJ, Fink LH, O’Hara B, Atwood WJ, Sullivan CS. Evolutionarily conserved function of a viral microRNA. J Virol. 2008;82:9823–8. doi:10.1128/JVI.01144-08.
Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–81. doi:10.1126/science.1113329).
Cui L, Guo X, Qi Y, Qi X, Ge Y, Shi Z, Wu T, Shan J, Shan Y, Zhu Z, Wang H. Identification of microRNAs involved in the host response to enterovirus 71 infection by a deep sequencing approach. J Biomed Biotechnol. 2010;2010:425939. doi:10.1155/2010/425939.
Qi Y, Tu J, Cui L, Guo X, Shi Z, Li S, Shi W, Shan Y, Ge Y, Shan J, Wang H, Lu Z. High-throughput sequencing of microRNAs in adenovirus type 3 infected human laryngeal epithelial cells. J Biomed Biotechnol. 2010;2010:915980. doi:10.1155/2010/915980.
Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–80. doi:10.1126/science.1102755).
Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–66. doi:10.1101/gad.1210204).
Van Stry M, Oguin TH, Cheloufi S, Vogel P, Watanabe M, Pillai MR, Dash P, Thomas PG, Hannon GJ, Bix M. Enhanced susceptibility of Ago1/3 double-null mice to influenza A virus infection. J Virol. 2012;86:4151–7. doi:10.1128/JVI.05303-11).
Singh A, Harris R. Autocrine, paracrine and juxtacrine signaling by EGFR ligands. Cell Signal. 2005;17:1183–93.
Belting M, Wittrup A. Nanotubes, exosomes, and nucleic acid-binding peptides provide novel mechanisms of intercellular communication in eukaryotic cells: implications in health and disease. J Cell Biol. 2008;183:1187–91. doi:10.1083/jcb.200810038).
Chen X, Liang H, Zhang J, Zen K, Zhang CY. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22:125–32. doi:10.1016/j.tcb.2011.12.001).
Simons M, Raposo G. Exosomes: vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–81. doi:10.1016/j.ceb.2009.03.007.
Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi:10.1038/ncb1596.
Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–6. doi:10.1038/ncb1800).
Zomer A, Vendrig T, Hopmans ES, van Eijndhoven M, Middeldorp JM, Pegtel DM. Exosomes: fit to deliver small RNA. Commun Integr Biol. 2010;3:447–50. doi:10.4161/cib.3.5.12339.
Pfeffer S, Zavolan M, Grässer FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. Identification of virus-encoded microRNAs. Science. 2004;304:734–6. doi:10.1126/science.1096781.
Choi H, Lee H, Kim SR, Gho YS, Lee SK. Epstein-Barr virus-encoded microRNA BART15-3p promotes cell apoptosis partially by targeting BRUCE. J Virol. 2013;87:8135–44. doi:10.1128/JVI.03159-12.
Ok CY, Papathomas TG, Medeiros LJ, Young KH. EBV-positive diffuse large B-cell lymphoma of the elderly. Blood. 2013;122:328–40. doi:10.1182/blood-2013-03-489708.
Skalsky RL, Corcoran DL, Gottwein E, Frank CL, Kang D, Hafner M, Nusbaum JD, Feederle R, Delecluse HJ, Luftig MA, Tuschl T, Ohler U, Cullen BR. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 2012;8:e1002484. doi:10.1371/journal.ppat.1002484.
Babu SG, Ponia SS, Kumar D, Saxena S. Cellular oncomiR orthologue in EBV oncogenesis. Comput Biol Med. 2011;41:891–8. doi:10.1016/j.compbiomed.2011.07.007.
Meckes DG, Shair KH, Marquitz AR, Kung CP, Edwards RH, Raab-Traub N. Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci USA. 2010;107:20370–5. doi:10.1073/pnas.1014194107.
Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Würdinger T, Middeldorp JM. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA. 2010;107:6328–33. doi:10.1073/pnas.0914843107.
Lazzeri E, Romagnani P. CXCR3-binding chemokines: novel multifunctional therapeutic targets. Curr Drug Targets Immune Endocr Metabol Disord. 2005;5:109–18.
Gourzones C, Gelin A, Bombik I, Klibi J, Vérillaud B, Guigay J, Lang P, Témam S, Schneider V, Amiel C, Baconnais S, Jimenez AS, Busson P. Extra-cellular release and blood diffusion of BART viral micro-RNAs produced by EBV-infected nasopharyngeal carcinoma cells. Virol J. 2010;7:271. doi:10.1186/1743-422X-7-271).
Author information
Authors and Affiliations
Corresponding author
Additional information
K. Okuyama, J. Ogata, N. Yamakawa contributed equally to this study.
About this article
Cite this article
Okuyama, K., Ogata, J., Yamakawa, N. et al. Small RNA as a regulator of hematopoietic development, immune response in infection and tumorigenesis. Int J Hematol 99, 553–560 (2014). https://doi.org/10.1007/s12185-014-1564-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12185-014-1564-4