Abstract
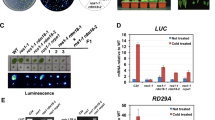
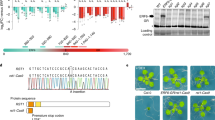
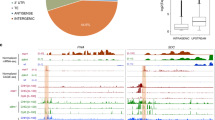
T-DNA insertion mutants have been widely used to investigate plant gene functions. Unexpectedly, in several reported cases, the phenotype of T-DNA insertion mutations can be suppressed because of trans T-DNA interactions associated with epigenetic modification, which indicates that caution is needed when T-DNA mutants are used. In the present study, we characterized a novel process suppressing a T-DNA mutation. The spz2 (suppressor of zou 2) mutant was isolated as a suppressor of the phenotype of the zou-4 mutant caused by a T-DNA insertion in the first intron. The spz2 mutation partially recovered the native ZOU gene expression in the zou-4 background, but not in two other zou alleles, zou-2 and zou-3, with T-DNAs inserted in the exon and intron, respectively. The suppressed phenotype was inherited in a Mendelian fashion and is not associated with epigenetic modification. The recovery of the native ZOU gene expression in the spz2 zou-4 double mutant is caused by transcriptional read-through of the intronic T-DNA as a result of decreased proximal polyadenylation. SPZ2 encodes an RNA-binding protein, FPA, which is known to regulate polyadenylation site selection. This is the first example of FPA rescuing a T-DNA insertion mutation by affecting the polyadenylation site selection.






Similar content being viewed by others
References
Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301:653–657
Bäurle I, Smith L, Baulcombe DC, Dean C (2007) Widespread role for the flowering-time regulators FCA and FPA in RNA-mediated chromatin silencing. Science 318:109–112
Chen S, Songkumarn P, Liu J, Wang GL (2009) A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol 150:1111–1121
Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17:1043–1054
Denay G, Creff A, Moussu S, Wagnon P, Thévenin J, Gérentes MF, Chambrier P, Dubreucq B, Ingram G (2014) Endosperm breakdown in Arabidopsis requires heterodimers of the basic helix-loop-helix proteins ZHOUPI and INDUCER OF CBP EXPRESSION 1. Development 141:1222–1227
Di Giammartino DC, Nishida K, Manley JL (2011) Mechanisms and consequences of alternative polyadenylation. Mol Cell 43:853–866
Duc C, Sherstnev A, Cole C, Barton GJ, Simpson GG (2013) Transcription termination and chimeric RNA formation controlled by Arabidopsis thaliana FPA. PLoS Genet 9:e1003867
Elkon R, Ugalde AP, Agami R (2013) Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet 14:496–506
Gan X, Stegle O, Behr J, Steffen JG, Drewe P, Hildebrand KL, Lyngsoe R, Schultheiss SJ, Osborne EJ, Sreedharan VT et al (2011) Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477:419–423
Gao Y, Zhao Y (2013) Epigenetic suppression of T-DNA insertion mutants in Arabidopsis. Mol Plant 6:539–545
Graber JH, Nazeer FI, Yeh PC, Kuehner JN, Borikar S, Hoskinson D, Moore CL (2013) DNA damage induces targeted, genome-wide variation of poly(A) sites in budding yeast. Genome Res 23:1690–1703
Hornyik C, Terzi LC, Simpson GG (2010) The spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev Cell 18:203–213
Ingram GC (2010) Family life at close quarters: communication and constraint in angiosperm seed development. Protoplasma 247:195–214
Jenal M, Elkon R, Loayza-Puch F, van Haaften G, Kuhn U, Menzies FM, Oude Vrielink JA, Bos AJ, Drost J, Rooijers K et al (2012) The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 149:538–553
Jia X, Chanda B, Zhao M, Brunner AM, Beers EP (2015) Instability of the Arabidopsis mutant csn5a-2 caused by epigenetic modification of intronic T-DNA. Plant Sci 238:53–63
Kanaoka MM, Pillitteri LJ, Fujii H, Yoshida Y, Bogenschutz NL, Takabayashi J, Zhu JK, Torii KU (2008) SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell 20:1775–1785
Kondou Y, Nakazawa M, Kawashima M, Ichikawa T, Yoshizumi T, Suzuki K, Ishikawa A, Koshi T, Matsui R, Muto S, Matsui M (2008) RETARDED GROWTH OF EMBRYO1, a new basic helix-loop-helix protein, expresses in endosperm to control embryo growth. Plant Physiol 147:1924–1935
Krysan PJ, Young JC, Sussman MR (1999) T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11:2283–2290
Lee BH, Henderson DA, Zhu JK (2005) The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17:3155–3175
Masamha CP, Xia Z, Yang J, Albrecht TR, Li M, Shyu AB, Li W, Wagner EJ (2014) CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature 510:412–416
Mayr C, Bartel DP (2009) Wide-spread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138:673–684
Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM (2007) SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19:1403–1414
Olsen OA (2004) Nuclear endosperm development in Cereals and Arabidopsis thaliana. Plant Cell 16:S214–S227
Oñate-Sánchez L, Vicente-Carbajosa J (2008) DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res Notes 1:93
Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53:247–259
Samson F, Brunaud V, Balzergue S, Dubreucq B, Lepiniec L, Pelletier G, Caboche M, Lecharny A (2002) FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res 30:94–97
Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB (2008) Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320:1643–1647
Sandhu KS, Koirala PS, Neff MM (2013) The ben1-1 brassinosteroid-catabolism mutation is unstable due to epigenetic modifications of the intronic T-DNA insertion. G3 3:1587–1595
Schomburg FM, Patton DA, Meinke DW, Amasino RM (2001) FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell 13:1427–1436
Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y (2011) Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA 17:761–772
Shi Y (2012) Alternative polyadenylation: new insights from global analyses. RNA 18:2105–2117
Shi Y, Manley JL (2015) The end of the message: multiple protein–RNA interactions define the mRNA polyadenylation site. Genes Dev 29:889–897
Spies N, Nielsen CB, Padgett RA, Burge CB (2009) Biased chromatin signatures around polyadenylation sites and exons. Mol Cell 36:245–254
Stangeland B, Salehian Z (2002) An improved clearing method for GUS assay in Arabidopsis endosperm and seeds. Plant Mol Bio Rep 20:107–114
Stroud H, Greenberg MVC, Feng S, Bernatavichute YV, Jacobsen SE (2013) Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152:352–364
Sussman MR, Amasino RM, Young JC, Krysan PJ, Austin-Phillips S (2000) The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol 124:1465–1467
Tanaka H, Onouchi H, Kondo M, Hara-Nishimura I, Nishimura M, Machida C, Machida Y (2001) A subtilisin-like serine protease is required for epidermal surface formation in Arabidopsis embryos and juvenile plants. Development 128:4681–4689
Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J (2007) JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signaling. Nature 448:661–665
Tian B, Manley JL (2013) Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem Sci 38:312–320
Ulker B, Peiter E, Dixon DP, Moffat C, Capper R, Bouché N, Edwards R, Sanders D, Knight H, Knight MR (2008) Getting the most out of publicly available T-DNA insertion lines. Plant J 56:665–677
Wang YH (2008) How effective is T-DNA insertional mutagenesis in Arabidopsis. J Biochem Technol 1:11–20
Xiang Y, Nakabayashi K, Ding J, He F, Bentsink L, Soppe WJJ (2014) Reduced Dormancy5 encodes a protein phosphatase 2C that is required for seed dormancy in Arabidopsis. Plant Cell 26:4362–4375
Xing Q, Creff A, Waters A, Tanaka H, Goodrich J, Ingram GC (2013) ZHOUPI controls embryonic cuticle formation via a signalling pathway involving the subtilisin protease ABNORMAL LEAFSHAPE1 and the receptor kinases GASSHO1 and GASSHO2. Development 140:770–779
Xue W, Ruprecht C, Street N, Hematy K, Chang C, Frommer WB, Persson S, Niittyla T (2012) Paramutation-like interaction of T-DNA loci in Arabidopsis. PLoS ONE 7:e51651
Yang S, Johnston N, Talideh E, Mitchell S, Jeffree C, Goodrich J, Ingram G (2008) The endosperm-specific ZHOUPI gene of Arabidopsis thaliana regulates endosperm breakdown and embryonic epidermal development. Development 135:3501–3509
Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L, Wang J (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34:293–297
Acknowledgments
This work was supported by the National Nature Science Foundation of China (grant nos. 31470286, 30970278 and 31271743) and the China National Transgenic Major Program (grant no. 2014ZX0800943B) and the Natural Science Foundation of Shandong Province of China (grant no. ZR2011CM020) and was also supported by the One Hundred Person Project of the Chinese Academy of Sciences.
Author contributions
S.X.Y., X.Z.F. and J.G. conceived the project and designed this work. Y.H.Z. and X.L. constructed the mutation library and performed map-based cloning. Y.H.Z., X.L. and C.X.W. performed transgenic, cell biological and other functional analyses. H.C.W. performed RNA sequencing analysis. S.X.Y., X.Z.F. and J.G. wrote this paper.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Yaohua Zhang and Xin Li contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, Y., Li, X., Goodrich, J. et al. Reduced function of the RNA-binding protein FPA rescues a T-DNA insertion mutant in the Arabidopsis ZHOUPI gene by promoting transcriptional read-through. Plant Mol Biol 91, 549–561 (2016). https://doi.org/10.1007/s11103-016-0487-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-016-0487-2