Abstract
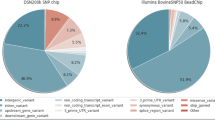
Cattle and water buffalo belong to the same subfamily Bovinae and share chromosome banding and gene order homology. In this study, we used genome-wide Illumina BovineSNP50 BeadChip to analyze 91 DNA samples from three breeds of water buffalo (Nili-Ravi, Murrah and their crossbred with local GuangXi buffalos in China), to demonstrate the genetic divergence between cattle and water buffalo through a large single nucleotide polymorphism (SNP) transferability study at the whole genome level, and performed association analysis of functional traits in water buffalo as well. A total of 40,766 (75.5 %) bovine SNPs were found in the water buffalo genome, but 49,936 (92.5 %) were with only one allele, and finally 935 were identified to be polymorphic and useful for association analysis in water buffalo. Therefore, the genome sequences of water buffalo and cattle shared a high level of homology but the polymorphic status of the bovine SNPs varied between these two species. The different patterns of mutations between species may associate with their phenotypic divergence due to genome evolution. Among 935 bovine SNPs, we identified a total of 9 and 7 SNPs significantly associated to fertility and milk production traits in water buffalo, respectively. However, more works in larger sample size are needed in future to verify these candidate SNPs for water buffalo.


Similar content being viewed by others
References
Fernandez MH, Vrba ES (2005) A complete estimate of the phylogenetic relationships in ruminantia: a dated species-level supertree of the extant ruminants. Biol Rev Camb Philos Soc 80:269–302
FAO (2000) Water buffalo: an asset undervalued. FAO Regional Office for Asia and Pacific, Bangkok
Vijh RK, Tantia MS, Mishra B, Bharani Kumar ST (2008) Genetic relationship and diversity analysis of Indian water buffalo (Bubalus bubalis). J Anim Sci 86:1495–1502
Kandasamy S, Jain A, Kumar R, Agarwal SK, Joshi P, Mitra A (2010) Molecular characterization and expression profile of uterine serpin (SERPINA14) during different reproductive phases in water buffalo (Bubalus bubalis). Anim Reprod Sci 122:33–141
Amaral ME, Grant JR, Riggs PK, Stafuzza NB, Filho EA, Goldammer T, Weikard R, Brunner RM, Kochan KJ, Greco AJ, Jeong J, Cai Z, Lin G, Prasad A, Kumar S, Saradhi GP, Mathew B, Kumar MA, Miziara MN, Mariani P, Caetano AR, Galvão SR, Tantia MS, Vijh RK, Mishra B, Kumar ST, Pelai VA, Santana AM, Fornitano LC, Jones BC, Tonhati H, Moore S, Stothard P, Womack JE (2008) A first generation whole genome RH map of the river buffalo with comparison to domestic cattle. BMC Genomics 9:631
Michelizzi VN, Dodson MV, Pan Z, Amaral ME, Michal JJ, McLean DJ, Womack JE, Jiang Z (2010) Water buffalo genome science comes of age. Int J Biol Sci 6:333–349
NCBI (2011) http://www.ncbi.nlm.nih.gov/sites/entrez
Matukumalli LK, Lawley CT, Schnabel RD, Taylor JF, Allan MF, Heaton MP, O’Connell J, Moore SS, Smith TP, Sonstegard TS, Van Tassell CP (2009) Development and characterization of a high density SNP genotyping assay for cattle. PLoS ONE 4:e5350
Decker JE, Pires JC, Conant GC, McKay SD, Heaton MP, Chen K, Cooper A, Vilkki J, Seabury CM, Caetano AR, Johnson GS, Brenneman RA, Hanotte O, Eggert LS, Wiener P, Kim JJ, Kim KS, Sonstegard TS, Van Tassell CP, Neibergs HL, McEwan JC, Brauning R, Coutinho LL, Babar ME, Wilson GA, McClure MC, Rolf MM, Kim J, Schnabel RD, Taylor JF (2009) Resolving the evolution of extant and extinct ruminants with high-throughput phylogenomics. Proc Natl Acad Sci USA 106:18644–18649
Aulchenko YS, Ripke S, Isaacs A, van Duijn CM (2007) GenABEL: an R library for genome-wide association analysis. Bioinformatics 23:1294–1296
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575
Kovach W (2005) MVSP-A multivariate statistical package for windows. Kovach Computing Services, Pentraeth
Pritchard J, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Gilmour AR, Gogel BJ, Cullis BR, Thompson R (2006) ASREML User Guide. Release 2.0. VSN International Ltd., Hatfield
Aulchenko YS, de Koning DJ, Haley C (2007) Genomewide rapid association using mixed model and regression: a fast and simple method for genomewide pedigree-based quantitative trait loci association analysis. Genetics 177:577–585
Amin N, van Duijn CM, Aulchenko YS (2007) A genomic background based method for association analysis in related individuals. PLoS ONE 2:e1274
Michelizzi VN, Wu X, Dodson MV, Michal JJ, Zambrano-Varon J, McLean DJ, Jiang Z (2011) A global view of 54,001 single nucleotide polymorphisms (SNPs) on the Illumina BovineSNP50 BeadChip and their transferability to water buffalo. Int J Biol Sci 7:18–27
Loftus RT, MacHugh DE, Bradley DG, Sharp PM, Cunningham P (1994) Evidence for two independent domestications of cattle. Proc Natl Acad Sci USA 91:2757–2761
Cockrill WR (1981) The water buffalo: a review. Br Vet J 137:8–16
Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R et al (2007) Genome-wide detection and characterization of positive selection in human populations. Nature 449:913–918
Streisfeld MA, Rausher MD (2010) Population genetics, pleiotropy, and the preferential fixation of mutations during adaptive evolution. Evolution 65:629–642
Ciaudo C, Servant N, Cognat V, Sarazin A, Kieffer E, Viville S, Colot V, Barillot E, Heard E, Voinnet O (2009) Highly dynamic and sex-specific expression of microRNAs during early ES cell differentiation. PLoS Genet 5:e1000620
Wu MY, Eldin KW, Beaudet AL (2008) Identification of chromatin remodeling genes Arid4a and Arid4b as leukemia suppressor genes. J Natl Cancer Inst 100:1247–1259
Chang G, Xu S, Dhir R, Chandran U, O’Keefe DS, Greenberg NM, Gingrich JR (2010) Hypoexpression and epigenetic regulation of candidate tumor suppressor gene CADM-2 in human prostate cancer. Clin Cancer Res 16:5390–5401
Allebrandt KV, Amin N, Müller-Myhsok B, Esko T et al (2011) A K(ATP) channel gene effect on sleep duration: from genome-wide association studies to function in Drosophila. Mol Psychiatry. doi: 10.1038/mp.2011.142
Lovasz N, Ducza E, Gaspar R, Falkay G (2011) Ontogeny of sulfonylurea-binding regulatory subunits of K(ATP) channels in the pregnant rat myometrium. Reproduction 142:175–181
Bryan J, Muñoz A, Zhang X, Düfer M, Drews G, Krippeit-Drews P, Aguilar-Bryan L (2007) ABCC8 and ABCC9: ABC transporters that regulate K+ channels. Pflugers Arch 453:703–718
Minoretti P, Falcone C, Aldeghi A, Olivieri V, Mori F, Emanuele E, Calcagnino M, Geroldi D (2006) A novel Val734Ile variant in the ABCC9 gene associated with myocardial infarction. Clin Chim Acta 370:124–128
Lu B, Jiang D, Wang P, Gao Y, Sun W, Xiao X, Li S, Jia X, Guo X, Zhang Q (2011) Replication study supports CTNND2 as a susceptibility gene for high myopia. Invest Ophthalmol Vis Sci 52:8258–8261
Ismail HM, Medhat AM, Karim AM, Zakhary NI (2011) FHIT gene and flanking region on chromosome 3p are subjected to extensive allelic loss in Egyptian breast cancer patients. Mol Carcinog 50:625–634
Syeed N, Husain SA, Sameer AS, Chowdhri NA, Siddiqi MA (2011) Mutational and promoter hypermethylation status of FHIT gene in breast cancer patients of Kashmir. Mutat Res 707:1–8
Acknowledgments
We thank the farm staff for helping to collect blood samples. Financial assistance from Chinese Projects from the Chinese government (201303118, 2011ZX08007-003, 31272427, 2011DFA32250, 2010-Z16, 2011-G26, YJH0044, 201070934339, 201171034317, 20110146110005), Innovation China UK (ICUK PoC-RVC-003), the National Modern Agro-industry Technology Research System of China (nycytx-10), and the earmarked fund for Modern Agro-industry Technology Research System (No. CARS-37-04B) are greatly appreciated.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2012_1932_MOESM3_ESM.doc
Supplementary Figure S1 The triangle plot of the three clusters using STRUCTURE 2.2 (CR: Crossbred; ML: Murrah; and NL: Nili-Ravi) (DOC 93 kb)
Rights and permissions
About this article
Cite this article
Wu, J.J., Song, L.J., Wu, F.J. et al. Investigation of transferability of BovineSNP50 BeadChip from cattle to water buffalo for genome wide association study. Mol Biol Rep 40, 743–750 (2013). https://doi.org/10.1007/s11033-012-1932-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-1932-1