Abstract
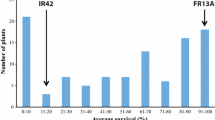
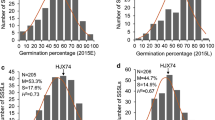
Submergence is a widespread problem of rice production, especially in low-lying areas in South and Southeast Asia. Despite the success of Sub1 mega varieties, repeated instances of prolonged and severe flooding in stress-prone areas suggests that the SUB1 gene is no longer sufficient in those regions and requires improved varieties with increased tolerance. A study was conducted to identify quantitative trait loci (QTLs) associated with submergence tolerance using 115 F7 recombinant inbred lines (RILs) derived from the cross of Ciherang-Sub1, a popular Indonesian cultivar carrying the SUB1 gene that has relatively higher tolerance to submergence compared to the performance of most other Sub1 lines and the submergence and stagnant flooding tolerant IR10F365. As the tolerant allele at SUB1A on chromosome 9 was fixed in this mapping population, additional QTLs responsible for submergence tolerance were expected to be revealed. Genotyping with an Infinium 6K SNP chip resulted in 469 polymorphic markers that were then used for QTL mapping. Phenotyping was performed under complete submergence with two replicates. A major QTL for submergence derived from Ciherang-Sub1, named qSUB8.1, was detected on chromosome 8 with a LOD score of 10.3 and phenotypic variance of 27.5%. Additionally, a smaller QTL, also derived from Ciherang-Sub1, was detected on chromosome 2 with a LOD score of 3.5 and phenotypic variance of 12.7%. There was no digenic interaction detected between these QTLs suggesting their independent action. The QTLs detected in this study can be used in marker-assisted selection to further improve the tolerance of other Sub1 varieties.


Similar content being viewed by others
References
Adkin SW, Shiraishi T, McComb JA (1990) Submergence tolerance of rice-a new glasshouse method for the experimental submergence of plants. Physiol Plant 80:642–646
Collard BCY, Septiningsih EM, Das SR, Carandang JJ, Pamplona AM, Sanchez DL, Kato Y, Ye G, Reddy JN, Singh US, Iftekharuddaula KM, Venuprasad R, Vera-Cruz CN, Mackill DJ, Ismail AM (2013a) Developing new flood-tolerant varieties at the International Rice Research Institute (IRRI). SABRAO J Breed Genet 45:42–56
Collard BCY, Kato Y, Septiningsih IAM, Mackill DJ (2013b) Defining IRRI’s role in the EIRLSBN: current status and future directions. In: Collard BCY, Ismail AM, Hardy B (eds) EIRLSBN: twenty years of achievements in rice breeding. International Rice Research Institute, Los Baños, pp 135–144
Dey MM, Upadhyaya HK (1996) Yield loss due to drought, cold and submeMappingrgence in Asia. In: Everson RE (ed) Rice research in Asia: progress and priorities. International Rice Resreach Institute, Manila, pp 291–303
Fukao T, Bailey-Serres J (2008) Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellins responses in rice. Proc Natl Acad Sci 105:16814–16819
Gonzaga ZJ, Carandang J, Sanchez DL, Mackill DJ, Septiningsih EM (2016) Mapping additional QTLs from FR13A to increase submergence tolerance beyond SUB1. Euphytica 209:627–636
Haque QA, Hille Ris Lambers D, Tepora NM, Dela Cruz QD (1989) Inheritance of submergence tolerance in rice. Euphytica 41:247–251
Herdt RW (1991) Research priorities for rice biotechnology. In: Khush GS, Toenniessen GH (eds) Rice biotechnology. CAB International, Oxon, pp 19–54
Iftekharuddaula KM, Newaz MA, Salam MA, Ahmed HU, Mahbub MAA, Septiningsih EM, Collard BCY, Sanchez DL, Pamplona AM, Mackill DJ (2011) Rapid and high-precision marker assisted backcrossing to introgress the SUB1 QTL into BR11, the rainfed lowland rice mega variety of Bangladesh. Euphytica 178:83–97
Iftekharuddaula KM, Salam MA, Newaz MA, Ahmed HU, Collard BCY, Septiningsih EM, Sanchez DL, Pamplona AM, Mackill DJ (2012) Comparison of phenotypic versus marker-assisted background selection for the SUB1 QTL during backcrossing in rice. Breed Sci 62:216–222
Iftekharuddaula KM, Ghosal S, Gonzaga ZJ, Amin A, Barman HN, Yameen R, Carandang J, Collard BYC, Septiningsih EM (2016a) Allelic diversity of newly characterized submergence-tolerant rice (Oryza sativa L.) germplasm from Bangladesh. Genet Resour Crop Evol 63:859–867
Iftekharuddaula KM, Ahmed HU, Ghosal S, Amin A, Moni ZR, Ray BP, Barman HN, Siddique MA, Collard BCY, Septiningsih EM (2016b) Development of early maturing submergence-tolerant rice varieties for Bangladesh. Field Crops Res 190:44–53
Joehanes R, Nelson JC (2008) QGene 4.0, an extensible java QTL-analysis platform. Bioinformatics 24:2788–2789
Kato Y, Collard BCY, Septiningsih EM, Ismail AM (2014) Physiological analyses of traits associated with tolerance of long-term partial submergence in rice. AoB PLANTS 6:plus058.
Khush GS (1984) Terminology of rice growing environments. International Rice Resreach Institute, Manila, pp 5–10
Li Z-X, Septiningsih EM, Quiloy-Mercado SM, McNally KL, Mackill DJ (2010) Identification of Sub1A alleles from wild rice Oryza rufipogon Griff. Genet Resour Crop Evol 58:1237–1242
Mackill DJ, Coffman WR, Garrity DP (1996) Rainfed lowland rice improvement. International Rice Research Institute, Los Baños
Manly KF, Cudmore RH Jr, Meer JM (2001) Map manager QTX, cross-platform software for genetic mapping. Mamm Genome 12:930–932
McCouch SR (2008) Gene nomenclature system for rice. Rice 1:72–84
Meng L, Li H, Zhang L, Wang J (2015) QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J 3:368–283
Mohanty HK, Khush GS (1985) Diallel analysis of submergence tolerance in rice, Oryza sativa L. Theor Appl Genet 70:467–473
Nandi S, Subudhi PK, Senadhira D, Manigbas NL, Sen-Mandi S, Huang N (1997) Mapping QTLs for submergence tolerance in rice by AFLP analysis and selective genotyping. Mol Gen Genet 255:1–8
Neeraja CN, Maghirang-Rodriquez R, Pamplona A, Heuer S, Collard BCY, Septiningsih EM, Vergara G, Sanchez D, Xu K, Ismail AM, Mackill DJ (2007) A marker-assisted backcross approach for developing submergence-tolerant rice cultivars. Theor Appl Genet 115:767–776
Septiningsih EM, Pamplona AM, Sanchez DL, Neeraja CN, Vergara GV, Heuer S, Ismail AM, Mackill DJ (2009) Development of submergence tolerant rice cultivars: the Sub1 locus and beyond. Ann Bot 103:151–160
Septiningsih EM, Sanchez DL, Singh N, Sendon PMD, Pamplona AM, Heuer S, Mackill DJ (2012) Identifying novel QTLs for submergence tolerance in rice cultivars IR72 and Madabaru. Theor Appl Genet 124:867–874
Septiningsih EM, Collard BCY, Heuer S, Bailey-Serres J, Ismail AM, Mackill DJ (2013) Applying genomics tools for breeding submergence tolerance in rice. In: Varshney RK, Tuberosa R (eds) Translational genomics for crop breeding: abiotic stress, yield and quality, 1st edn. John Wiley and Sons, Chichester, pp 9–30
Septiningsih EM, Hidayatun N, Sanchez DL, Nugraha Y, Carandang J, Pamplona AM, Collard BCY, Ismail AM, Mackill DJ (2015) Accelerating the development of new submergence tolerant rice varieties: the case of Ciherang-Sub1 and PSBRc18-Sub1. Euphytica 202:259–268
Setter TL, Ellis M, Laureles EV, Ella ES, Senadhira D, Mishra SB, Sarkarung S, Datta S (1997) Physiology and genetics of submergence tolerance in rice. Ann Bot 79:67–77
Siangliw M, Toojinda T, Tragoonrung S, Vanavichit A (2003) Thai jasmine rice carrying QTLchr9 (SubQTL) is submergence tolerant. Ann Bot 91:255–261
Singh N, Dang T, Vergara G, Pandey D, Sanchez D, Neeraja C, Septiningsih E, Mendioro M, Teeson-Mendoza R, Ismail A, Mackill D, Heuer S (2010) Molecular marker survey and expression analyses of the rice submergence-tolerance genes SUB1A and SUB1C. Theor Appl Genet 121:1441–1453
Singh R, Singh Y, Xalaxo S, Verulkar S, Yadav N, Singh S, Singh N, Prasad KSN, Kondayya K, Rao PVR, Rani MG, Anuradha T, Suraynarayana Y, Sharma PC, Krishnamurthy SL, Sharma SK, Dwivedi JL, Singh AK, Singh PK, Nilanjay SNK, Kumar R, Chetia SK, Ahmad T, Rai M, Perraju P, Pande A, Singh DN, Mandal NP, Reddy JN, Singh ON, Katara JL, Marandi B, Swain P, Sarkara RK, Singh DP, Mohapatra T, Padmawathi G, Ram T, Kathiresan RM, Paramsivam K, Nadarajan S, Thirumeni S, Nagarajan M, Singh AK, Vikram P, Kumar A, Septiningsih E, Singh US, Ismail AM, Mackill D, Singh NK (2016) From QTL to variety-harnessing the benefits of QTLs for drought, flood, and salt tolerance in mega varieties of India through a multi-institutional network. Plant Sci 242:278–287
Sinha MM, Saran S (1988) Inheritance of submergence tolerance in lowland rice. Oryza 25:351–354
Suprihatno B, Coffman WR (1981) Inheritance of submergence tolerance in rice (Oryza sativa L.). SABRAO J 13:98–108
Thomson MJ (2014) High-throughput SNP genotyping to accelerate crop improvement. Plant Breed Biotech 2:195–212
Toojinda T, Siangliw M, Tragroonrung S, Vanavichit A (2003) Molecular genetics of submergence tolerance in rice: QTL analysis of key traits. Ann Bot 91:243–253
Vergara BS, Mazaredo A (1975) Screening for resistance to submergence under greenhouse conditions. In: Proceedings International Seminar on Deepwater Rice. Bangladesh Rice Research Institute, Dhaka, Bangladesh pp 67–70
Wang S, Basten CJ, Zeng Z-B (2010) Windows QTL Cartographer 2.5. Department of Statistics. North Carolina State University, Raleigh
Xu K, Mackill DJ (1996) A major locus for submergence tolerance mapped on rice chromosome 9. Mol Breed 2:219–224
Xu K, Xu X, Ronald PC, Mackill DJ (2000) A high-resolution linkage map of the vicinity of the rice submergence tolerance locus Sub1. Mol Gen Genet 263:681–689
Xu K, Xia X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ (2006) Sub1A is an ethylene response factor-like gene that confers submergence tolerance to rice. Nature 442:705–708
Yang J, Zhu J (2005) Methods for predicting superior genotypes under multiple environments based on QTL effects. Theor Appl Genet 110:1268–1274
Yang J, Zhu J, Williams RW (2007) Mapping the genetic architecture of complex traits in experimental populations. Bioinformatics 23:1527–1536
Yang J, Hu CC, Hu H, Yu RD, Xia Z, Ye XZ, Zhu J (2008) QTL network: mapping and visualizing genetic architecture of complex traits in experimental populations. Bioinformatics 24:721–723
Zheng K, Subudhi PK, Domingo J, Magpantay G, Huang N (1995) Rapid DNA isolation for marker assisted selection in rice breeding. Rice Genet Newslett 12:255–258
Acknowledgements
We thank R. Garcia, E. Suiton, J. Mendoza, G. Perez, J. Borgonia, R. Formaran, V. Bartolome and the IRRI GSL team for technical assistance. The work reported here was supported in part by a grant from the Bill and Melinda Gates Foundation (BMGF) through the project on “Stress-tolerant rice for Africa and South Asia (STRASA)”, Global Rice Science Partnership (GRiSP), and the National Institute of Food and Agriculture, U.S. Department of Agriculture, Hatch project 1009300.
Authors’ contributions
E.M.S. designed the experiments. B.C.Y.C developed the RIL population using rapid generation advance (RGA). J.C. performed the seed multiplication of the mapping population. Z.J.C.G and A.S. performed the DNA extraction, and J.C. and Z.J.C.G performed the phenotyping under the supervision of E.M.S. M.J.T. supervised the SNP genotyping in the Genotyping Service Lab (GSL). Z.J.C.G., A.S., and E.M.S. analyzed the data. Z.J.C.G. and E.M.S. wrote the manuscript. B.C.Y.C. and M.J.T. edited the manuscript. All authors read and approved the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Electronic supplementary material
Supplementary Table 1
Recombinant Inbred Lines (RILs) with average survival rates above the tolerant check FR13A containing combinations of the identified QTLs (90–100% survival rates). (DOCX 13 kb)
Supplementary Figure 1
A representative figure showing the QTLs for submergence tolerance from Ciherang-sub1/IR10F365 population identified by QTLNetwork. The red balls indicate the QTLs with additive effects. (DOCX 49 kb)
Rights and permissions
About this article
Cite this article
Gonzaga, Z.J.C., Carandang, J., Singh, A. et al. Mapping QTLs for submergence tolerance in rice using a population fixed for SUB1A tolerant allele. Mol Breeding 37, 47 (2017). https://doi.org/10.1007/s11032-017-0637-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-017-0637-5