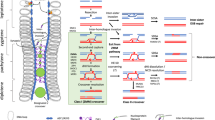
Abstract
The low frequency of meiotic recombination in chromosomal regions other than hotspots is a general obstacle to efficient breeding. A number of active genes are present in recombination-repressed centromeric regions in higher eukaryotes, suggesting that suppression of meiotic recombination prevents shuffling of genes within a centromeric region. In this study, by using an inter-subspecific cross of Oryza sativa L., we show that modification of inactive chromatin states by either genetic or chemical inhibition of chromatin modifying proteins induced changes in both the position of meiotic recombination and, unexpectedly, the pattern of segregation distortion of parental alleles. Antisense knockdown of rice homologues of DECREASE IN DNA METHYLATION1, which is required for the maintenance of heterochromatin in Arabidopsis thaliana, induced a recombination hotspot in a centromeric region accompanied by a steep increase in the proportion of heterozygotes. Our results describe a previously undocumented phenomenon in which artificial chromatin modification could be used to change the pattern of segregation distortion in rice and open up novel possibilities for efficient crop breeding.



Similar content being viewed by others
References
Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, Jacobsen SE (2008) Genome-wide association of histone H3 lysine nine methylation with CHG methylation in Arabidopsis thaliana. PLoS ONE 3:e3156
Black BE, Bassett EA (2008) The histone variant CENP-A and centromere specification. Curr Opin Cell Biol 20:91–100
Bühler M, Gasser SM (2009) Silent chromatin at the middle and ends: lessons from yeasts. EMBO J 28:2149–2161
Burt A, Trivers R (2006) Genes in conflict. The biology of selfish genetic elements. Belknap Press, Cambridge
Colomé-Tatché M, Cortijo S, Wardenaar R et al (2012) Features of the Arabidopsis recombination landscape resulting from the combined loss of sequence variation and DNA methylation. Proc Natl Acad Sci USA 109:16240–16245
Copenhaver GP, Nickel K, Kuromori T et al (1999) Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286:2468–2474
Folco HD, Pidoux AL Urano T, Allshire RC (2008) Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science 319:94–97
Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Tagami Y, Nakayama T, Oshimura M (2004) Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol 6:784–791
Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA (2002) Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297:1871–1873
Harushima Y, Kurata N, Yano M, Nagamura Y, Sasaki T, Minobe Y, Nakagahra M (1996) Detection of segregation distortions in an indica-japonica rice cross using a high-resolution molecular map. Theor Appl Genet 92:145–150
Higo H, Tahir M, Takashima K et al (2012) DDM1 (Decrease in DNA Methylation) genes in rice (Oryza sativa). Mol Genet Genomics 287:785–792
Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133:523–536
Lyttle TW (1991) Segregation distorters. Annu Rev Genet 25:511–557
Martinez-Peres E, Moore G (2008) To check or not check? The application of meiotic studies to plant breeding. Curr Opin Plant Biol 11:222–227
Melamed-Bessudo C, Levy AA (2012) Deficiency in DNA methylation increases meiotic crossover rates in euchromatic but not in heterochromatic regions in Arabidopsis. Proc Natl Acad Sci USA 109:E981–E988
Mirouze M, Lieberman-Lazarovich M, Aversano R, Bucher E, Nicolet J, Reinders J, Paszkowski J (2012) Loss of DNA methylation affects the recombination landscape in Arabidopsis. Proc Natl Acad Sci USA 109:5880–5885
Nagaki K, Cheng Z, Ouyang S, Talbert PB, Kim M, Jones KM, Henikoff S, Buell CR, Jiang J (2004) Sequencing of a rice centromere uncovers active genes. Nat Genet 36:138–145
Numa H, Kim JM, Matsui A et al (2010) Transduction of RNA-directed DNA methylation signals to repressive histone marks in Arabidopsis thaliana. EMBO J 29:352–362
Perrella G, Consiglio MF, Aiese-Cigliano R, Cremona G, Sanchez-Moran E, Barra L, Errico A, Bressan RA, Franklin FC, Conicella C (2010) Histone hyperacetylation affects meiotic recombination and chromosome segregation in Arabidopsis. Plant J 62:796–806
Rhoades MM (1942) Preferential segregation in maize. Genetics 27:395–407
Saffery R, Sumer H, Hassan S, Wong LH, Craig JM, Todoroki K, Anderson M, Stafford A, Choo KHA (2003) Transcription within a functional human centromere. Mol Cell 12:509–516
Satoh K, Doi K, Nagata T et al (2007) Gene organization in rice revealed by full-length cDNA mapping and gene expression analysis through microarray. PLoS ONE 2:e1235
Talbert PB, Henikoff S (2010) Centromeres convert but don’t cross. PLoS Biol 8:e1000326
Vongs A, Kakutani T, Martienssen RA, Richards EJ (1993) Arabidopsis thaliana DNA methylation mutants. Science 260:1926–1928
Wijnker E, de Jong H (2008) Managing meiotic recombination in plant breeding. Trends Plant Sci 13:640–646
Wu C-I, Lyttle TW, Wu M-L, Lin G-F (1988) Association between a satellite DNA sequence and the Responder of Segregation Distorter in D. melanogaster. Cell 54:179–189
Wu J, Mizuno H, Hayashi-Tsugane M et al (2003) Physical maps and recombination frequency of six rice chromosomes. Plant J 36:720–730
Yan H, Jin W, Nagaki K, Tian S, Ouyang S, Buell CR, Talbert PB, Henikoff S, Jiang J (2005) Transcription and histone modifications in the recombination-free region spanning a rice centromere. Plant Cell 17:3227–3238
Yan H, Talbert PB, Lee HR, Jett J, Henikoff S, Chen F, Jiang J (2008) Intergenic location of rice centromeric chromatin. PLoS Biol 6:e286
Yelina NE, Choi K, Chelysheva L et al (2012) Epigenetic remodeling of meiotic crossover frequency in Arabidopsis thaliana DNA methyltransferase mutants. PLoS Genet 8:e1002844
Acknowledgments
We thank K.-I. Nonomua for the RCS2 centromeric probe; T. Kanno and H. Rothnie for their comments on this study; K. Hioki, H. Onodera and A. Tagiri for technical assistance. This work was supported by grants of Promotion of Research Activity and NIAS Strategic Research Fund from National Institute of Agrobiological Sciences, NIBB Cooperative Research Program (Next-generation DNA Sequencing Initiative: 11-703), the Ministry of Agriculture, Forestry and Fisheries (PGE1004), and JST/CREST, Japan.
Conflict of interest
National Institute of Agrobiological Sciences holds a patent on genetic and chemical modification of positions of meiotic recombination described in this paper.
Author information
Authors and Affiliations
Corresponding author
Additional information
Accession codes The genome re-sequencing data have been deposited in the DNA Data Bank of Japan under the accession number DRX001982, and the microarray data have been deposited in NCBI Gene Expression Omnibus under accession number GSE31775.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Habu, Y., Ando, T., Ito, S. et al. Epigenomic modification in rice controls meiotic recombination and segregation distortion. Mol Breeding 35, 103 (2015). https://doi.org/10.1007/s11032-015-0299-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-015-0299-0