Abstract
Much of the vast literature on time series classification makes several assumptions about data and the algorithm’s eventual deployment that are almost certainly unwarranted. For example, many research efforts assume that the beginning and ending points of the pattern of interest can be correctly identified, during both the training phase and later deployment. Another example is the common assumption that queries will be made at a constant rate that is known ahead of time, thus computational resources can be exactly budgeted. In this work, we argue that these assumptions are unjustified, and this has in many cases led to unwarranted optimism about the performance of the proposed algorithms. As we shall show, the task of correctly extracting individual gait cycles, heartbeats, gestures, behaviors, etc., is generally much more difficult than the task of actually classifying those patterns. Likewise, gesture classification systems deployed on a device such as Google Glass may issue queries at frequencies that range over an order of magnitude, making it difficult to plan computational resources. We propose to mitigate these problems by introducing an alignment-free time series classification framework. The framework requires only very weakly annotated data, such as “in this ten minutes of data, we see mostly normal heartbeats\(\ldots \),” and by generalizing the classic machine learning idea of data editing to streaming/continuous data, allows us to build robust, fast and accurate anytime classifiers. We demonstrate on several diverse real-world problems that beyond removing unwarranted assumptions and requiring essentially no human intervention, our framework is both extremely fast and significantly more accurate than current state-of-the-art approaches.
Similar content being viewed by others
1 Introduction
Time series classification has been a highly active research area in the last decade, with at least several hundred published papers on this topic appearing in the literature (Batista et al. 2011; Bao and Intille 2004; Chen et al. 2005; Hu et al. 2013, 2011; Keogh et al. 2006, 2004a, b; Morse and Patel 2007; Niennattrakul et al. 2010; Rakthanmanon et al. 2011; Ratanamahatana and Keogh 2004; Ueno et al. 2010). The majority of research efforts test the utility of their ideas on the UCR classification archive, which with forty-seven time series datasets, is the largest such benchmark in the world (Keogh et al. 2006). We believe that the availability of this (admittedly very useful) resource has isolated much of the research community from the realities of deploying classifiers in the real world, and as a consequence, many claims for the utility of time series classification algorithms have unwarranted optimism. The problem is that the UCR data is formatted in a canonical way that makes comparisons between rival methods easy, but isolates researchers from certain problems that are likely to reduce the performance of all algorithms when deployed in realistic settings, by an amount that dwarfs the relatively minor differences between rival approaches. Below we consider the four issues we have identified as creating a gulf between most academic work and practical deployable systems.
In all the time series in the UCR classification archive, the time series has been processed into short equal-length “template” sequences that are representative of the class. For example, individual and complete gait cycles for biometric classification (Andino 2000; Faezipour et al. 2010; Hanson et al. 2009; Koch et al. 2010), individual and complete heartbeats for cardiological classification (de Chazal et al. 2004; Hu et al. 2011), individual and complete gestures for gesture recognition (Yang et al. 2009), etc.
In most cases, the segmentation of long time series into these idealized snippets is done by hand (Hanson et al. 2009; Koch et al. 2010; Lester et al. 2005; www.mocap.cs.cmu.edu/). However, for many real-world problems this either cannot be done, or only done with great effort (Gafurov and Snekkenes 2008; Liu et al. 2010; Pärkkä et al. 2006).
As a concrete example, consider the famous Gun/Point problem (Keogh et al. 2006; Ratanamahatana and Keogh 2004), which has appeared in well over one hundred works (Chen et al. 2005; Keogh et al. 2004a; Morse and Patel 2007). To create this dataset, the original authors (Ratanamahatana 2012; Ratanamahatana and Keogh 2004) used a metronome that signaled every three seconds to cue both the actor’s behavior and the start/stop of the recording apparatus (Ratanamahatana 2012). This allowed the extraction of perfectly aligned data, containing all of the target behavior and only the target behavior. Unsurprisingly, dozens of papers report less than 10 % classification error rate on this problem. However, it seems unlikely that this error rate reflect our abilities with real-world data.
Such contriving of time series datasets appears to be the norm in the literature. For example, (Yang et al. 2009) notes, “one subject performed one trial of an action (in exactly) 10 s.” and (www.pamap.org/demo.html) tells us that human editors should carefully discard “ all transient activities between performing different activities.” Likewise, a recent paper states: “We assume that the trajectories are segmented in time such that the first and last frames are already aligned (and) the resulting model has the same length” (Usabiaga et al. 2007). Another recent paper that considers motion capture data tells us “While the original motion sequences have different lengths, we trim them with equal duration” (Li and Aditya 2011). Furthermore, the location of this trimming is subjective, relying on the ability to find the region “most significant in telling human motion apart, (as) suggested by domain experts” (Li and Aditya 2011). Note that these authors are to be commended for stating their assumptions so concretely. In the vast majority of cases, no such statements are made, however it is all but certain that similar “massaging” of the data has occurred.
We believe that such contriving of the data has led to unwarranted optimism about how well we can classify real-time series data streams. For real-world problems, we cannot always expect the training data to be so idealized, and we certainly cannot expect the testing data to be so perfect.
Moreover, in virtually all time series classification research, the data must be arranged to have equal length (Usabiaga et al. 2007). For example, in the world’s largest collection of time series datasets, the UCR classification archive, all forty-seven time series datasets contain only equal-length data (Keogh et al. 2006).
An additional assumption made in the vast majority of the literature is that all objects to be classified belong to exactly one of two or more well-defined classes. For example, in the Gun/Point problem, every one of the instances is either a gun-aiming or a finger-pointing (unarmed) behavior. However, the vast majority of normal human actions are clearly neither. How well do current techniques work when most of the data is not from the well-defined classes? A handful of papers consider this issue at testing time, using a rejection threshold to define the other class. However, to the best of our knowledge, this issue has not been considered at training time. Instead, as noted above, it is simply assumed that some mechanism is able to provide perfect training data. With a handful of exceptions (i.e. some types of heartbeat extraction Yang et al. 2012), this “mechanism” is costly and subjective human labor. A more realistic idea for data gathering is to capture data “in the wild” as in Bao and Intille (2004), Pham et al. (2010), Reiss and Stricker (2011), etc. However, this opens the problem of data editing and cleaning. For example, a 1-h trace of data labeled “walking” will almost certainly contain non-representative subsequences, such as the subject pausing at a crosswalk, or introducing a temporary asymmetry into her gait as she answers her phone. The current solution to preprocess such data requires human intervention to examine and edit such traces, and keeping data that demonstrate the sought-after variability (walking uphill, downhill, level, walking fast, normal, slow), while discarding data that is atypical of the class.
The fourth and final unrealistic assumption that is common in the literature is that queries to be classified are presented to the classification system at equal time intervals. For example, if we know a system will produce queries ten times a second, we can then plan the hardware resources needed, and the maximum size of the training set. However, in many real world systems the available time for classification is not known a priori and may change as a consequence of external circumstances (Shieh and Keogh 2010). For example, for some ECG classification systems, the individual beats are detected, and then passed to the classification system. Given that human heart rates vary from about 40 to 200 beats per minute, the query arrival rate can range between 0.6 and 3.3 Hz.Footnote 1 Another example that recently came to our attention involves insect classification (Hao et al. 2013). The classification of flying insects can be fruitfully considered a time series problem and there the arrival rates can vary by at least four orders of magnitude (Batista et al. 2011; Hao et al. 2013). If we plan only for the fastest possible arrival rate, then we may be forced to invest in computational resources that are unused 99.99 % of the time, or to only consider a tiny training dataset, when 99.99 % of the time we could have availed of the larger dataset, and achieved a higher accuracy.
To summarize, much of the progress in time series classification from streams in the last decade is almost certainly optimistic, given that most of the literature implicitly or explicitly assumes one or more of the following:
-
1.
Copious amounts of perfectly aligned atomic patterns can be obtained (Hanson et al. 2009; Vatavu 2011; Yang et al. 2009).
-
2.
The patterns are all of equal length (Hanson et al. 2009; Keogh et al. 2006; Koch et al. 2010; Pärkkä et al. 2006; Reiss and Stricker 2011).
-
3.
Every item that we attempt to classify belongs to exactly one of our well-defined classes. Moreover, even at training time we have access to data that belongs to exactly one class (Gafurov and Snekkenes 2008; Keogh et al. 2006; Pärkkä et al. 2006; Ratanamahatana and Keogh 2004).
-
4.
The queries arrive at a constant rate that is known ahead of time (Kranen and Seidl 2009; Ueno et al. 2010).
In this work, we demonstrate a time series classification framework that does not make any of these assumptions. This lack of assumptions means our system can be deployed with much less human effort, and can significantly outperform other approaches, which often relied on subjective choices in training set creation.
Our approach requires only very weakly-labeled data, such as “This ten-minute trace of ECG data consists mostly of arrhythmias, and that three-minute trace seems mostly free of them”, removing assumption (1). Using this data we automatically build a “data dictionary”, which contains only the minimal subset of the original data to span the concept space. This is because the data dictionary can contain, say, one example of walking fast, one example of walking normal, etc. This mitigates assumption (2).
As a byproduct of building this data dictionary, we learn a rejection threshold, which allows us to address assumption (3). A query item further than this threshold to its nearest neighbor is assumed to be in the other class. We show that using the Uniform Scaling distance measure (Keogh et al. 2004b) instead of Euclidean distance also addresses assumption (2). Finally, we introduce a novel technique to search the data dictionary in an anytime manner (Shieh and Keogh 2010), allowing us to handle dynamic arrival rates and addressing assumption (4).
The rest of this paper is organized as follows: In Sect. 2, we introduce definitions and notation used in this paper. In Sect. 3.1, we show how classification is achieved given our data dictionary model. In Sect. 3.2, we illustrate how to actually learn the data dictionary by utilizing data editing techniques (Niennattrakul et al. 2010; Rakthanmanon et al. 2011; Ueno et al. 2010; Ye et al. 2009). Section 3.3 demonstrates how our framework learns the threshold distances. We demonstrate how we remove the fourth assumption by using the algorithm introduced in Sect. 3.4. In Sect. 4, we present a detailed empirical evaluation of our ideas. We discuss related work in Sect. 5. Finally, in Sect. 6, we offer conclusions and directions for future work.
2 Definitions and notation
We begin by defining all the necessary notation, starting with the definition of the data type of interest, time series:
Definition 1
(Time series) \(\hbox {T} = t_{1},{\ldots }, t_{m}\) is an ordered set of m real-valued variables.
We are only interested in local properties of a time series, thus we confine our interest to subsequences:
Definition 2
(Subsequence) Given a time series T of length m, a subsequence \(\hbox {S}_{k}\) of T is a sampling of length \(\hbox {n} \le \hbox {m}\) of contiguous position from T with starting position at \(k,\,\hbox {S}_{k} = \hbox {t}_{k},{\ldots },\hbox {t}_{k+n-1}\) for \(1 \le k \le m-n+1\).
The extraction of subsequences from a time series can be achieved by use of a sliding window:
Definition 3
(Sliding window) Given a time series T of length m, and a user-defined subsequence length of n, all possible subsequences can be extracted by sliding a window of size n across T and extracting each subsequence, \(S_{k}\). For a time series T with length m, the number of all possible subsequences of length n is \(m-n+1\).
For concreteness, we take the step of explicitly defining training data, as our definition of training data explicitly removes the assumptions inherent in most works (Hanson et al. 2009; Keogh et al. 2006; Koch et al. 2010; Pärkkä et al. 2006; Reiss and Stricker 2011; Usabiaga et al. 2007; www.mocap.cs.cmu.edu/).
Definition 4
(Training data) A Training Data C is a collection of the weakly-labeled time series annotated by behavior/state or some other mapping to the ground truth.
By weakly-labeled we simply mean that each long data sequence has a single global label and not lots of local labeled pointers to every beginning and ending of individual patterns, e.g., individual gestures. There are two important properties of such data that we must consider:
-
Weakly-labeled training data may contain extraneous/irrelevant sections. For example, after a subject reaches down to turn on an ankle sensor to record her gait, there may be a few seconds before she actually begins to walk (Reiss and Stricker 2011). Moreover, during the recording session, the subject may pause to shop, or jump to avoid a puddle. It seems very unlikely that such recordings could avoid having such spurious data. Note that this claim is not mere speculation; we observed this phenomenon in the first few seconds of the BIDMC Congestive Heart Failure dataset (www.physionet.org/physiobank/database/chfdb/) as shown in Fig. 1, and similar phenomena occur in all the datasets we examined.
-
Weakly-labeled training data will almost certainly contain significant redundancies. While we want lots of data in order to learn the inherent variability of the concept we wish to learn, significant redundancy will make our classification algorithms slow when deployed. Consider Fig. 1 once more. Once we have a single normal heartbeat, say pattern
 , then there is little utility in adding any of the 14 or so other very similar patterns, including pattern
, then there is little utility in adding any of the 14 or so other very similar patterns, including pattern  . However, to robustly learn this concept (beats belonging to Record-08), we must add either example of the Premature Ventricular Contraction (PVC).
. However, to robustly learn this concept (beats belonging to Record-08), we must add either example of the Premature Ventricular Contraction (PVC).
A snippet of BIDMC congestive heart failure database ECG—Record-08 (www.physionet.org/physiobank/database/chfdb/). a Is weakly-labeled data, which exhibits both extraneous data, a section of recording when the machine was not plugged in, and redundant data (only one pair of redundancies are shown in bold ( ). b A minimally redundant set of representative heartbeats (a data dictionary) could be used as training data (Color figure online)
). b A minimally redundant set of representative heartbeats (a data dictionary) could be used as training data (Color figure online)
Rather than these large weakly-labeled training datasets, we desire a smaller “smart” training data subset that does not contain spurious data, while maintaining coverage of the target concept by having one (ideally, exactly one) instance of each of the many ways the targeted behavior is manifest. For example, from the training data shown in Fig. 1, we want just one PVC example and just one example of a normal heartbeat (perhaps either
 or
or
 ). However, we do not want to require costly human effort to obtain this. While the time series shown in Fig. 1 would be fairly easy to edit by hand, it is only 0.16% of the full ECG dataset we consider in Sect. 4.2. Therefore, our objective is to build this idealized subset of the training data automatically. We begin by defining it more concretely as a data dictionary.
). However, we do not want to require costly human effort to obtain this. While the time series shown in Fig. 1 would be fairly easy to edit by hand, it is only 0.16% of the full ECG dataset we consider in Sect. 4.2. Therefore, our objective is to build this idealized subset of the training data automatically. We begin by defining it more concretely as a data dictionary.
Definition 5
A Data Dictionary D is a (potentially very small) “smart” subset of the training data. We allow an input parameter x, where x is the percentage of the training data C used in data dictionary D. The range of x is (0,100 %], and a dictionary with the percentage x of the original data is denoted as \(\mathbf{D}_{{{\mathbf {x}}}}\).
As the Data Dictionary is at the heart of our contribution, we will take the time to discuss it in detail.
2.1 A discussion of data dictionaries
As defined above, there are a huge number of possible data dictionaries for any percentage x, as any random subset of C satisfies the definition. However, we obviously wish to create one with some desirable properties.
Clearly, the classification error rate obtained from using just D should be no worse than that obtained from using all the training data. We do not wish to sacrifice accuracy. As we shall show, this is a surprisingly easy objective to achieve. In fact, as we shall show later, the classification error rate using a judiciously chosen D is generally significantly lower than using all of C. This is because the data dictionary contains less spurious -and therefore, potentially misleading-data.
Another very desirable property of D is that it be a very small percentage of the training data. This is to allow real-time deployment of the classifier, especially on resource limited devices (embedded devices, smartphones, etc. Bao and Intille 2004; Gafurov et al. 2006). This requirement may be seen as conflicting with the above classification error rate requirement; however, again we will show that in three diverse real-world problems we can judiciously throw away more than 95 % of C to obtain a \(\mathbf{D}_{\mathbf{5}{\varvec{\%}} }\) that is at least as accurate as using all the data in C.
Note that the number of subsequences within each class in D may be different. That is to say, our algorithm for building D is not round-robin; rather the algorithm adaptively adds more subsequences to cover the more “complicated” classes of D. For example, the ECG data from Record-08 shown in Fig. 1 is relatively simple. In contrast, the ECG of Record-03 shown in Fig. 2 has a more complicated trace, and at least four kinds of beats (normal, S, PVC and Q). Therefore, we might expect the number of subsequences for Record-03 in D to be greater than that for Record-08, something that is empirically borne out in our experiments (Sect. 4).
A snippet of BIDMC Congestive Heart Failure Database ECG: Record-03 (www.physionet.org/physiobank/database/chfdb/). Note that this section of ECG data exhibits more variability than the data in Fig. 1
Finally, there is the question of what value we should set x to. In fact, we can largely bypass this issue by providing an algorithm that produces a “spectrum” of data dictionaries in the range of \(x= (0,100~\%]\), together with an estimate of their error rate on unseen data. The user can examine this error rate vs. value-of-x curve to make the necessary trade-offs. Note that these data dictionaries are “nested”, that is to say, for any value of x we have \(\mathbf{D}_{{{\mathbf {x}}}} \subseteq \mathbf{D}_{{{\mathbf {x}}}+{\varvec{\varepsilon }}}\). Thus, we can consider our data dictionary creation algorithm an anyspace algorithm Ye et al. (2009).
Given the above considerations, how can we build the best data dictionary? As we will later show, we can heuristically search the space of data dictionaries using the simple algorithm in Sect. 3.2.
2.2 An additional insight on data redundancy
Based on our experience with real-world time series problems, we noted the following: in many cases, D contains many patterns that appear to be simply (linearly) rescaled versions of each other. For clarity, we illustrate our point with a synthetic example in Fig. 3; however, we will later show some real examples.
This situation is a consequence of our requirement that data dictionary D has the most representative subsequences of training data C. For example, if one class contains examples of walk, we hope to have at least one representative of each type of walk—perhaps one example of a leisurely-amble, one example of a normal-paced-walk, one example of a brisk-walk, etc. It is important to note that in this example, the three walking styles are not simply linearly rescaled versions of each other. They have different foot strike patterns, and thus produce different prototypical time series templates (Cavagna et al. 1977; McMahon and Cheng 1990). Nevertheless, within each sub-class of walk, there may also be a need to allow some linear rescaling of the time series. Using the Euclidean distance our search algorithm can achieve this by attempting to ensure that the data dictionary contains each gait pattern over a range of speeds. This is what our toy example in Fig. 3 illustrates.
Left A toy example data dictionary which was condensed from a large dataset. These seven subsequences in data dictionary A span the concept space of the bulls/bears problem. Right Note that if we had a distance measure that was invariant to linear scaling, we could further reduce data dictionary A to data dictionary B
For example, when reducing a dataset of daily human activities, we may have to extract examples of a brisk- walk at \(6.0, 6.1, 6.2\,\hbox {km/h}\), etc. However, by generalizing from the Euclidean distance to the Uniform Scaling distance (Keogh et al. 2004b), we allow our algorithm to keep just one example of the walk, and still achieve coverage of the target concept by using a flexible measure instead of lots of data. The Uniform Scaling distance is a simple generalization of the Euclidean distance that allows limited invariance of the length of the patterns being matched (Keogh et al. 2004b). The maximum amount of linear scaling allowed is a user-defined parameter (Keogh et al. 2004b). As we later show, allowing just a small amount of scaling, say 25 %, can greatly improve accuracy.
To see this in a real dataset, consider Fig. 4 left, which shows one of 15 classes that was processed into a data dictionary in an experiment we performed in Sect. 4.2. At first glance, the two patterns seem redundant,Footnote 2 violating one of the requirements stated above.
Left A data dictionary learned from a 15-class ECG classification problem (just class 01 is shown here). At first glance, the two exemplars seem redundant apart from their (irrelevant) phases. Right By using the Euclidean distance between the two patterns we can see that the misalignment of the beats would cause a large error. The problem solved by using the Uniform Scaling distance (Keogh et al. 2004b)
Instead of having two similar but different scaled patterns, just a single pattern is kept using the Uniform Scaling distance. We have found that using the Uniform Scaling distance allows us to have a significantly smaller data dictionary. In Fig. 4, we could delete either one of the two patterns and cover the space of possible heartbeats from Record-01. For example, in Fig. 3, we could further delete patterns I, II and IV and still cover the space of possible “bulls”.
However, beyond reducing the size of data dictionaries (thus speeding up classification), there is an additional advantage of using Uniform Scaling; it allows us to achieve a lower error rate. How is this possible? It is possible because we can generalize to patterns not seen in the training data.
Imagine the training data does contain some examples of gaits at speeds from 6.1 to \(6.5\,\hbox {km/h}\). As noted above, if the data dictionary has enough examples to cover this range of speeds, we should expect to do well. However, suppose the unseen data contains some walking at \(6.7\,\hbox {km/h}\). This is only slightly faster than we have seen in the training data, but the Euclidean distance is very sensitive to such changes (Keogh et al. 2004b). Using the Uniform Scaling distance allows us to generalize our labeled example at \(6.5\,\hbox {km/h}\) to the brisker \(6.7\,\hbox {km/h}\) instance. This idea is more than speculation. As we show in Sect. 4, using the Uniform Scaling distance does produce a significantly lower error rate.
2.3 On the need for a rejection threshold
As noted above, the training set may have extraneous data. Likewise, in most realistic deployment scenarios, we expect some (often most) of the data to be classified as the other class. In these cases, we wish our algorithm to label the objects as such. To achieve this, the data dictionary must have a distance threshold r beyond which we reject the query as unclassifiable (i.e., the other class). As we will show, we can learn this threshold as we build the dictionary.
3 Algorithms
In order to best explain our framework, we first assume a data dictionary with the appropriate threshold has already been created and begin by explaining how our classification model works. Later, in Sect. 3.2, we revisit the more difficult task of learning the data dictionary.
3.1 Classification using a data dictionary
Our classification model requires just a data dictionary with its accompanying threshold distance, r.
For an incoming object to be classified q, we classify it with the data dictionary using the classic nearest neighbor algorithm (Ueno et al. 2010). In Table 1, we show how to determine the class membership of this query, including the possibility that this query does not belong to any class in this data dictionary. For our purposes, there are exactly two possibilities of interest:
If the query’s nearest neighbor distance is larger than the threshold distance, we say this query does not belong to any class in this data dictionary (line 12).
If the query’s nearest neighbor distance is smaller than the threshold distance, then it is assigned to the same class as its nearest neighbor (line 14).
The algorithm begins by initializing the bsf distance to infinity and the predicted class_label to NaN in lines 1 and 2. From lines 3 to 9, we find the nearest neighbor of the query q in data dictionary D. The subroutine NN_search (shown in Table 2) returns the nearest neighbor distance of q within a time series. If the nearest neighbor distance within a time series in line 4 is smaller than the bsf, then in lines 6 and 7 we update the bsf and the class_label.
From lines 11 to 15, we compare the nearest neighbor distance to the threshold distance r. If the nearest neighbor distance is smaller than r, then this query belongs to the same class as its nearest neighbor. Otherwise, this query does not belong to any class within this data dictionary and is thus classified as the other class.
As we show in Table 1 line 4, the function NN_search is slightly different from the classic nearest neighbor search algorithm (Keogh et al. 2006). NN_search returns not only the nearest neighbor distance of a query, but also a distance vector that contains distances between the query and all the possible subsequences in a time series. This distance vector is not exploited at classification time, but as we show in Sect. 3.2, it is exploited when building the data dictionary. For concreteness, we briefly discuss the NN_search function in Table 2 below.
In line 1, using a sliding window (cf. Definition 3), we extract all the subsequences of the same length as the query. From lines 3 to 5, the distances between q and all the possible subsequences are calculated. We calculate the nearest neighbor distance in line 6. Note that in line 4, the distance could be Euclidean distance (Keogh et al. 2006), or Uniform Scaling distance (Keogh et al. 2004b), etc. We will revisit this choice in Sect. 4.
In addition to finding the nearest neighbor, this function also returns a distance vector. This additional information is exploited by the dictionary building algorithm discussed later in Sect. 3.2. Figure 5 bottom shows an example of such a distance vector.
Top A snippet of BIDMC Congestive Heart Failure Database ECG data: Record-08 (www.physionet.org/physiobank/database/chfdb/). Bottom the distance vector of an incoming query. The nearest neighbor and its distance of q is colored in  (Color figure online)
(Color figure online)
Having demonstrated how the classification model works in conjunction with the data dictionary, we are in position to illustrate how to build the data dictionary, which is a more difficult task.
3.2 Building the data dictionary
As discussed in Sect. 2, we want to build the data dictionary automatically. Using human effort to manually edit the training data into a data dictionary is clearly not a realistic solution: as it is not scalable to large datasets and invites human bias into the process.
Before introducing our dictionary-building algorithm, we will show a worked example on a toy dataset in the discrete domain. We use a small discrete domain example simply because is it easy to write intuitively; our real goal remains large real-valued time series data.
3.2.1 The intuition behind data dictionary building
Suppose we have a training dataset that contains two classes, \(\hbox {C}_{1}\) and \(\hbox {C}_{2}\):

In this toy example, the data is weakly-labeled. The colored/ bolded text is for the reader’s introspection only; it is not available to the algorithm. Here the reader can see that in \(\hbox {C}_{1}\), there appears to be two ways a shorter subsequence query might belong to this class; if it contains the word  or
or  . This is similar to the situation shown in Fig. 1 where a query will be classified to the class of Record-08 if it contains pattern
. This is similar to the situation shown in Fig. 1 where a query will be classified to the class of Record-08 if it contains pattern  or pattern PVC.
or pattern PVC.
We want to know whether any incoming queries belong to either class in this training data or not. In our proposed framework, we search just the data dictionary.
Recall that one of the desired properties of the data dictionary is that it contains a minimally redundant set of patterns that is representative of the training data. In this example for \(\hbox {C}_{1}\), these are clearly the substrings  and
and  . Likewise for \(\hbox {C}_{2}\),
. Likewise for \(\hbox {C}_{2}\),  and
and  seem to completely define the class. Thus, the data dictionary D should be the following:
seem to completely define the class. Thus, the data dictionary D should be the following:

Consider now two incoming queries ieap and kklp. The former is a noisy version of a pattern found in our dictionary, but as it is within our rejection threshold of (hamming) distance r of 1, it is correctly labeled as \(\hbox {C}_{2}\). In contrast, kklp has a distance of 3 to its nearest neighbor in D, so it is correctly rejected.
Note that had we attempted to classify against the raw data rather than the dictionary, the query kklp would have been classified as \(\hbox {C}_{1}\) (it appears in the middle of  ). This misclassification is clearly contrived, but it does happen frequently in the real data. Consider the flat section of time series at the beginning of Fig. 1. As noted above, it is extraneous data, due to a temporary disconnection of the sensor. However, many other patients’ ECG traces also have these flat sections, but clearly that does not mean we should classify them as belonging to patient Record-08.
). This misclassification is clearly contrived, but it does happen frequently in the real data. Consider the flat section of time series at the beginning of Fig. 1. As noted above, it is extraneous data, due to a temporary disconnection of the sensor. However, many other patients’ ECG traces also have these flat sections, but clearly that does not mean we should classify them as belonging to patient Record-08.
In our example, we have considered two separate queries; however, a closer analogue of our real-valued problem is to imagine an endless stream that needs to be classified:
Up to this point we have not explained how we built our toy dictionary. The answer is simply to use the results of leaving-one-out classification to score candidate substrings (leaving-one-out classification is just a particular case of leave-p-out cross-validation with \(p = 1\)). For example, by using leaving-one-out to classify the first substring of length 4 in \(\hbox {C}_{1}\)
dpac, it is incorrectly classified as \(\hbox {C}_{2}\) (it matches the middle of  with a distance of 1). In contrast, when we attempt to classify the second substring of length 4 in \(\hbox {C}_{1}\), pace, we find it is correctly classified. By collecting statistics about which substrings are often used for correct predictions, but rarely used for wrong predictions, we find that the four substrings shown in our data dictionary emerge as the obvious choices. This basic idea is known as data editing (Niennattrakul et al. 2010; Pekalska et al. 2006; Ueno et al. 2010). In the next section, we formalize this idea, and generalize it to real-valued data streams.
with a distance of 1). In contrast, when we attempt to classify the second substring of length 4 in \(\hbox {C}_{1}\), pace, we find it is correctly classified. By collecting statistics about which substrings are often used for correct predictions, but rarely used for wrong predictions, we find that the four substrings shown in our data dictionary emerge as the obvious choices. This basic idea is known as data editing (Niennattrakul et al. 2010; Pekalska et al. 2006; Ueno et al. 2010). In the next section, we formalize this idea, and generalize it to real-valued data streams.
3.2.2 Building the data dictionary
The high-level intuition behind building the data dictionary is to use a ranking function to score every subsequence in C. These “scores” rate the subsequences by their expected utility for classification of future unseen data. We use these scores to guide a greedy search algorithm, which iteratively selects the best subsequence and places it in D. How do we know this utility? We simply estimate it by cross validation, e.g. looking at the classification error rate and some additional information as explained below.
As previously hinted, our algorithm iteratively adds subsequences to the data dictionary. Each iteration has three steps. In Step 1, the algorithm scores the subsequences in C. In Step 2, the highest scoring subsequence is extracted and placed in D. Finally, in Step 3, we identify all the queries that cannot be correctly classified by the current D. These incorrectly classified items are passed back to Step 1 to re-score the subsequences in C.
There is an important caveat. Once we have removed the best subsequence in Step 2, the scores of all the other subsequences may change in the next iteration. To return to our running example in Fig. 1, either subsequence  and
and  would rank highly. However once we have placed one, say
would rank highly. However once we have placed one, say  , in D, there is little utility in adding
, in D, there is little utility in adding  , since having
, since having  in D is sufficient to correctly classify similar patterns in Step 3. Thus we expect the scores of
in D is sufficient to correctly classify similar patterns in Step 3. Thus we expect the scores of  will be low in the next iteration, given that the correctly classified queries by the current D will not be used to re-score C in the next iteration.
will be low in the next iteration, given that the correctly classified queries by the current D will not be used to re-score C in the next iteration.
The process iterates until we run out of subsequences to add to D or the unlikely event of perfect training error rate having been achieved. In the dozens of problems we have considered, the training error rate plateaus well before 10 % of the training data has been added to the data dictionary.
Below we consider each step in detail.
Step 1 In order to rank every point in the time series, we use the leaving-one-out classification algorithm.Footnote 3 However, we do not want to use just the classification error rate to score the subsequences. Imagine we have two subsequences \(\hbox {S}_{1}\) and \(\hbox {S}_{2}\), either of which is found to correctly predict 70 % of the queries tested with them. Either appears to be a good candidate to add to D. However, suppose that in addition to being close enough to many objects with the same class label (friends), allowing its 30 % error rate, further suppose that \(\hbox {S}_{1}\) is also very close to many objects with different class labels (enemies). If \(\hbox {S}_{2}\) keeps a larger distance from its enemy class objects, it is a much better choice for inclusion in D.
This idea, that instead of using just the error rate of classification, you must also consider the relative distance to “friends” and “enemies” has been investigated extensively in the field of data editing (Pekalska et al. 2006; Ueno et al. 2010).
Given a query length l, we randomly choose a query q from the training data C.Footnote 4 In Table 3, lines 2 and 3, we first split the training data into two parts, Part A (friends only) and Part B (enemies only). Using the NN_search algorithm in Table 2, we find nearest neighbor
friend in Part A (lines  –
– ) and nearest neighbor
enemy (lines
) and nearest neighbor
enemy (lines  –
– ) in Part B.
) in Part B.
In lines 23–27, the nearest neighbor friend distance and the nearest neighbor enemy distance are compared. If the nearest neighbor friend distance is smaller than the nearest neighbor enemy distance, we discover all the distances of the query q in Part A that are also smaller than the nearest neighbor enemy distance. Such subsequences are likely true positives. That is to say, our confidence that these subsequences can produce correct classifications of unseen data has increased.
Similarly, if the nearest neighbor friend distance is larger than the nearest neighbor enemy distance, we find all the distances of the query q in Part B that are also smaller than the nearest neighbor friend distance. We call the corresponding subsequences likely false positives.
 , the blue text describes
, the blue text describes  )
)Given the likely true/false positives found in Table 3, we are now in a position to discuss how to rank them. The algorithm to prioritize their inclusion in the training set will use this ranking.
By utilizing the simple rank function introduced in Ueno et al. (2010), we generalize an algorithm that gives positive score to likely true positives and negative score to the likely false positives.
Note that subsequences that are not used to classify any queries (correctly or not) get a zero score. Using a large number of queries, we compute a score vector for every time series in C. We denote rank(S) as the score for a subsequence S in the time series.
In the next step, we demonstrate how to extract the current best subsequence using the score vectors.
Step 2 We extract the highest scoring subsequence and place it in D. We demonstrate this step by using the example in Fig. 6. Suppose in one of the iterations in Step 1, the starting point of the  /bold heartbeat has the highest score. Therefore we need to extract this heartbeat. Because the Euclidean distance is very sensitive to even slight misalignments and our scoring function is somewhat “blurred” as to its exact location in the x-axis. Extracting exactly the subsequence with query length l would be very brittle. Therefore, we “pad” the chosen subsequence some time series from the left and to the right, in particular with the l / 2 data points to either side.
/bold heartbeat has the highest score. Therefore we need to extract this heartbeat. Because the Euclidean distance is very sensitive to even slight misalignments and our scoring function is somewhat “blurred” as to its exact location in the x-axis. Extracting exactly the subsequence with query length l would be very brittle. Therefore, we “pad” the chosen subsequence some time series from the left and to the right, in particular with the l / 2 data points to either side.
Top A snippet of BIDMC Congestive Heart Failure Database ECG data: Record-08 (www.physionet.org/physiobank/database/chfdb/). Bottom the extracted subsequence has twice the query length
Note that there is a slight difference between the first iteration and the subsequent iterations. Before the first iteration, D is empty. After the first iteration, D should contain exactly one subsequence from each class. This is the smallest D logically possible. Therefore, instead of splitting C to the friends part and the enemies part, the algorithm finds the most representative subsequence in each class in Step 1, and then adds them into D in Step 2.
After the first iteration, we extract only the one subsequence that holds the highest score in C and add it into D. Thus, the class sizes in D can be skewed, as the algorithm adds more exemplars to the more diverse/complicated classes. While we are iteratively building D, the size of C becomes smaller, as the extracted subsequence is removed from C.
Step 3 The algorithm examines the quality of the current D by doing classification using all the queries. The queries that are correctly classified by the current D will not be used to re-score C in the next iteration Step 1, since the current D is sufficient to correctly classify them. Only the misclassified queries will proceed back to Step 1 to re-score C. In each iteration Step 3, we redo classification experiments on D using all the queries, since the correctly classified queries in \(\mathbf{D}_{{{\mathbf {x}}}}\) may become misclassified in \(\mathbf{D}_{{{\mathbf {x}}}+{\varvec{\varepsilon }}}\).
After building a data dictionary for a training data, our last obligation is to learn the distance threshold.
3.3 Learning the threshold distance
After the data dictionary is built, we learn a threshold to allow us to reject future queries, which do not belong to any of our learned classes. We begin by recording a histogram of the nearest neighbor distances of testing queries that are correctly classified using D, as shown in Fig. 7. Next, we compute a similar histogram for the nearest neighbor distances of queries, which should not have a valid and meaningful match within D (i.e., the other class). Where can we get such queries? In the example shown in Fig. 7, we simply used gesture data as the other class, knowing gestures should not match a set of heartbeats. Note that it is occasionally possible that a gesture will match a heartbeat by coincidence; but our approach is robust to such spurious matches so long as they are relatively rare. If external datasets are in short supply, we can also simply permute subsequences of D to produce the other class, for example flipping heartbeats upside-down and backwards.
The  left histogram contains the nearest neighbor distances of correctly classified queries for the ECG data used in Sect. 4.2. The
left histogram contains the nearest neighbor distances of correctly classified queries for the ECG data used in Sect. 4.2. The  /right histogram shows nearest neighbor distances for queries from the other class (Color figure online)
/right histogram shows nearest neighbor distances for queries from the other class (Color figure online)
Given the two histograms, we choose the location that gives the equal-error-rate as the threshold (about 7.1 in Fig. 7). However, based on their tolerance to false negatives, users may choose a more liberal or conservative decision boundary.
3.4 Anytime classification
For many time series problems, the available time to classify an instance may vary over many orders of magnitude. As a concrete example, consider the task of flying insect classification, which (Hao et al. 2013) recently suggested can be fruitfully considered a time series problem. As Fig. 8 shows, the arrival rates of the insects can vary by at least four orders of magnitude (Batista et al. (2011); Hao et al. (2013)). While handling such a huge range is currently at the limit of what we can do, in Sect. 4.4 we show that we can address the order of magnitude range that is common in medicine (ECGs etc) and human behavior.
Culex. stigmatosoma is the mosquito which is the primary vector of Western Equine Encephalitis. We measured the arrival rate of male Cx. stigmatosoma at a sensor Hao et al. (2013) that can unobtrusively count them. The arrival rates are heavily correlated with day/night cycles, and differ over four orders of magnitude
This variability in time available to classify an instance is particularly problematic for a lazy learner such as nearest neighbor classification. If we plan only for the fastest possible arrival rate, then we will be forced to invest in computational resources that are unused 99.9 % of the time. Otherwise, we may consider a tiny training dataset, when 99.9 % of the time we could have availed of the larger dataset, and achieved a higher accuracy. To mitigate this issue we can cast our framework as an anytime algorithm, as explained in the next section.
3.4.1 Anytime classification
Anytime classification algorithms are algorithms that can sacrifice the quality of results (here accuracy) for a faster running time (Grass and Zilberstein 1995; Shieh and Keogh 2010; Zilberstein and Russell 1995). The algorithm becomes interruptible after a short time of initialization, then as shown in Fig. 9 offers a tradeoff between the quality of experimental result and the computation time.
An anytime classification algorithm can mitigate the assumption that the arriving rate of queries is known ahead of time, since the computation can be interrupted any time after a short period of initialization. For example, suppose that the Data Dictionary D contains 1000 exemplars, we begin to search the nearest neighbor for a query through all the 1000 exemplars. If the computation is not interrupted during this search, we can report the class label of the nearest exemplar as the predicted class (i.e Table 1). However, if the computation is interrupted before seeing all the 1000 exemplars, we can instead simply report the class label of the nearest exemplar with the bsf distance as the predicted class. Table 4 formalizes this intuition. The algorithm begins by initializing the bsf distance to infinity and the predicted class_label to NaN in lines 1 and 2. In line 3, we extracted all the possible subsequences in the training data in a specific order. Note that the order of all the subsequences can be defined by any arbitrary methods. For example, in Table 2 line 1, all possible subsequences are extracted in a left-to-right order using a sliding window. Thus the search process in Table 2 is essentially sequential search. Likewise, we could search in random order to make the algorithm invariant to some arbitrary order of data collection etc. To approximately maximize the desirable diminishing returns property of anytime algorithms, in Sect. 3.4.2, we propose to index all the possible subsequences using a novel technique.
In Table 4 line 4, we start to calculate the distance between q and each subsequence. If the distance in line 6 is smaller than the bsf, then in lines 7 and 8 we update the bsf and the class_label accordingly. From lines 10 to 12, if the stopFlag is true, then the computation will be terminated and return the current class_label associated with the bsf distance.
3.4.2 Using complexity to order the anytime search
In Table 4 line 3, we noted that the subsequences in the data dictionary can be sorted in any order. It is easy to see that some orders can result in very poor performance. For example, suppose we have a balanced two-class problem with 1,000 exemplars. If the data is sorted such that all the training exemplars belonging to class-1 are searched first, then any interruption before we have seen more than 500 objects will result in the algorithm predicting class-1. Either random ordering or round robin ordering can easily fix this pathological problem. However, this invites us to consider if there is an optimal ordering? In Ueno et al. (2010) the author argue that in the face of unknown objects to classify that the idea of an optimal ordering is ill-defined, however we can still (as they do) attempt to find a good ordering.
We propose to use the complexity of each subsequence as the ordering key. The idea is to compute the complexity of all objects in the data dictionary (this needs to be done just once, offline during the dictionary creation). When the query arrives, we measure its complexity, and then search the data dictionary in order to find the training exemplar that has the closest complexity with this query. The intuition is that if two sequences have very different complexities, then they must surely also have very high Euclidean distances (note the converse need not be true).
The complexity of a time series can be calculated by different methods, such as Kolmogorov complexity (Li and Vitanyi 1997), variants of entropy (Andino 2000; Aziz and Arif 2006), etc. There are several desirable properties of a complexity measure (Batista et al. 2011), such as,
-
Low time and space complexity;
-
Few parameters, ideally none;
-
Intuitive and interpretable;
Given the above consideration, we propose to use one complexity measure shown in Eq. (2), which has O(1) space and O(n) time complexity (Batista et al. 2011). More importantly, this complexity measure has a natural interpretation and no parameters.
We are not claiming this is the optimal ordering technique. We simply want to show an existence proof of an ordering technique that can mitigate the assumption (4). Table 5 outlines the algorithm. In line 1, we calculate the complexity of the incoming query. Line 2 shows that we extract all the possible subsequences in the data dictionary using a sliding window. The sliding window length is the same length as the query. This extraction is the same as the one in line 1 in Table 2. From lines 3 to 6, we calculate the absolute difference of the complexity between the query and each subsequence. Last, we sort the differences in an ascending order in line 7. After the sorting, the closer the subsequence is to the query in terms of complexity, the higher rank it will have. In other words, in Table 4 line 3, the subsequence that has the most similar complexity to the query, will have the highest priority for calculating its Euclidian distance with that query.
Before moving on, we will preempt a possible misunderstanding. The ordering calculated in Table 3, orders the data for our anyspace technique. If you can only afford to use \(D\%\) of the full dataset, the first \(D\%\) of the ordered data is what you should take. This ordering is computed exactly once, during training time.
In contrast, the ordering discussed in this section is used at testing time. Given a query, if we want to find its class label quickly, we should compare it to its nearest neighbor in each class. However finding the nearest neighbor requires a nearest neighbor similarity search. One could do this search in random order, but our claim is that if we do it in the complexity-based order, we are likely to find a “near enough” neighbor more quickly. In Sect. 4.4, we show that this ranking technique is very effective.
3.5 The utility of the uniform scaling technique
Finally, we can trivially replace the Euclidean distance with Uniform Scaling Footnote 5 distance in the above data dictionary building and threshold learning process (Keogh et al. 2004b). We choose the maximum scaling factor based on the variability of time series in the domain at hand, see discussion in Sect. 4. A naive implementation of Uniform Scaling would be slow, but Keogh et al. (2004b) shows that it can be computed in essentially the same time as Euclidean distance.
4 Experimental evaluation
We begin by discussing our experimental philosophy. To ensure that our experiments are easily reproducible, we have built a website, which contains all the datasets and code (https://sites.google.com/site/dmkdrealistic/). In addition, this website contains additional experiments, which are omitted here for brevity. Our experimental results support our claim that using only the data dictionary is more accurate and faster than using all the available training data. In Table 6, we summarize the datasets consider in this work.
We compare our algorithm with several widely used rival approaches. The most widely used rival approach extracts feature vectors from the data and reports the best result among multiple models (Bao and Intille 2004; Pham et al. 2010; Reiss and Stricker 2011). In addition, we compare with the obvious strawman of using all the training data, which is just a special case of our framework, in which all the training data is used (i.e. \(\mathbf{D}_{{\varvec{100\%}} })\).
To support our claim that the real-world streaming data is not as clean as the contrived datasets used in most literature, we report the percentage of the rejected queries produced by the learned threshold and show some examples.Footnote 6
We report the error rate using both Euclidean distance and Uniform Scaling distance to support our claim that the latter can be very useful for time series classification problems.
While we are ultimately interested in the testing error rate, we also report the training error rate, as this can be used to predict the best size of the data dictionary for a given problem. However, for completeness, we build and test the data dictionary \(\mathbf{D}_{{{\mathbf {x}}}}\) for every value of x, from the smallest logically possible size to whatever value minimizes the holdout error rate (this is generally much less than \(x = 10\,\%\)).
The reader may object that error rate is not the correct measure here. Imagine that our rejection threshold is so high that we reject 999 of 1000 queries, and just happen to get one classified object correct. In this case, reporting a 0 % error rate would be dubious at best. This is of course what precision/recall and similar measurements are designed to be robust to. However, in all our case studies, our rejection rate is much less than 10 %, so reporting just the error rate is reasonable, and allows us to present more visually intuitive figures. Moreover, we will show experiments where we consider the correctness of rejections made by our algorithm.
Finally, we defer experiments that consider the scalability of dictionary building to https://sites.google.com/site/dmkdrealistic/, noting in passing that this is done offline, and that in any case we can do this faster than real-time. In other words, we can learn the dictionary for an hour heartbeats in much less than one hour.
4.1 An example application in physiology
We consider a physical activity dataset called ‘PAMAP’, which contains eight subjects performing activities such as: normal-walking, walking-very-slow, descending-stairs, cycling, and inactivity (an umbrella term for lying-in-bed/sitting-still/standing-still), etc (www.pamap.org/demo.html). Approximately eight hours of data at 110 Hz was collected from wearable sensors on the subjects’ wrist, chest, and shoes.
The classification error rates for D from \(\hbox {D}_{0.39\%}\) to \(\hbox {D}_{14.2\%}\) for the physical activity dataset (www.pamap.org/demo.html)
For simplicity of exposition, we consider only a single time series, recording the roll-axis from the sensor placed in the subjects’ shoe. However, our algorithm trivially extends to multi-dimensional data (examples appear at https://sites.google.com/site/dmkdrealistic/). Note that although our algorithm only uses a single axis from the sensor, we demonstrate that our results are significantly better than rival algorithms that use all three-axis data (roll, pitch and yaw) from the same sensor (Reiss and Stricker 2011).
We randomly choose 60 % of the data as training data, and treat the rest as testing data. We repeat this randomly sampling of training and testing data twenty times and in Fig. 10 we show the average results over all this twenty training and testing sets. In Fig. 10, we show the training/testing error rates as our algorithm grows D from the smallest logically possible size (about 0.39 % of all the training data) to the point where it is clear that our algorithm can no longer improve. Although our algorithm bottoms out earlier in the plot, we wish to demonstrate that the output is very smooth over a wide range of values.
We compare with the widely-used rival approach (Bao and Intille 2004; Koch et al. 2010; Reiss and Stricker 2011), which extracts signal features from the sliding windows. For fairness to this method, we used their suggested window size Reiss and Stricker (2011), and tested all of the following classifiers: K-nearest neighbors (K = 5), SVM, Naïve Bayes, boosted decision trees and C4.5 decision tree (Bao and Intille 2004; Pham et al. 2010; Reiss and Stricker 2011). The best classification result is 0.364 achieved by the C4.5 decision tree.
For the commonly used strawman of using all the training data, the testing error rate is 0.221. However, our framework equals this testing error rate using only 1.6 % (i.e. \(\mathbf{D}_{{\varvec{1.6\%}}}\)) of the training data and obtains the significantly lower error rate of 0.152 at \(\mathbf{D}_{{\varvec{8.3\%}}}\) . Moreover, given that we are using only about one-twelfth the data, we are able to classify the data about twelve times faster.
Our algorithm is clearly highly competitive, but does it owe its performance to choice of which subsequences are placed in D by our algorithm? To test this, we built another D by randomly extracting subsequences from C. As Fig. 10 also shows, our systematic method for ranking subsequences is significantly better than random selection.
A final observation about these results is that the training error rate tends to be a very good predictor of the test error rate. As Fig. 10 shows, the training error is only slightly optimistic.
We are now ready to test our claim that Uniform Scaling (c.f. Sect. 2.2) can help in datasets containing signals acquired from human behavior/physiology. We repeated the experiments above under the exact same conditions, except we replaced Euclidean distance with Uniform Scaling distance in both the training and testing phases.
Based on studies of variability for human locomotion (Aspelin 2005; Cavagna et al. 1977; McMahon and Cheng 1990), we chose a maximum scaling factor of 15 %; that is to say, queries are tested at every scale from 85 to 115 % of their original length. Uniform Scaling obtains a 0.085 testing error rate at \(\mathbf{D}_{{\varvec{8.1\%}}}\), significantly better than Euclidean distance, as shown in Fig. 11.
We learned a threshold distance of 14.5 for D.Footnote 7 With this threshold, our algorithm rejects 9.5 % of the testing queries. In Fig. 12, we see that the vast majority of rejected queries do belong to the other class and are thus correctly rejected.
We do not present formal numerical results for the rejected queries, as the weakly- annotated format of the original data does not provide the label of the objects with certainty. Approximately 9.5 % of the queries where rejected, we carefully looked at a random sample of 100, and we are very confident they are true rejections.
This dataset draws from sporting activities. In Sect. 4.3 we also consider a similar but independent dataset, call the MIT benchmark Bao and Intille (2004), which additionally considers more quotidian activities such as tooth-brushing etc. We achieve near identical improvements on this dataset, giving us some confidence that the results here are not a happy coincidence.
Finally, we consider how much the size of the training data affects our algorithm. To do this we repeated the experiment shows in Fig. 10, which used 60 % of the all the data for training, but with progressively less training data. As shown in Fig. 13 using a smaller pool of data from which to build the classifier does hurt somewhat, but surprisingly little. Even if the training data is one-third the original size (just 20 % instead of 60 %) our algorithm can still learn a classifier that, while using only about 5 % of the data, is better than the strawman of using all the training data (the horizontal line at 0.221 in Fig. 13).
The experiment considered in Fig. 10, repeated for increasing smaller training set sizes
4.2 An example application in cardiology
We apply our framework to a large ECG dataset: the BIDMC Congestive Heart Failure Database (www.physionet.org/physiobank/database/chfdb/). The dataset includes ECG recordings from fifteen subjects with severe congestive heart failure. The individual recordings are each about 20 h in duration, sampled at 250 Hz.
Ultimately, the medical community wants to classify patient-independent types of heartbeats. However, in this experiment, we classify individuals’ heartbeats. This is simply because we are able to obtain huge amounts of labeled data this way. Note that as hinted at in Fig. 2, the data is complex and noisy. Moreover, a single (unhealthy) individual may have many different types of beats. Cardiologist Helga Van Herle from USC informs us this is a perfect proxy problem.
We use a randomly selected 150 min of data for training, and 450 min of data for testing.
In Fig. 14, we show the training/testing error rates as our algorithm grows the data dictionary from the smallest possible size \((\mathbf{D}_{{\varvec{0.28\%}} })\) to the point where it is clear that our algorithm can no longer improve.
Note that the testing error rate is 0.102 using the strawman of using all the training data, which is significantly better than the default error rate 0.933. However, our framework duplicates this error rate using only 2.1 % (i.e. \(\mathbf{D}_{{\varvec{2.1\%}}}\)) of the training data, and obtains the much lower error rate of 0.076 at \(\mathbf{D}_{{\varvec{4.5\%}}}\). From Fig. 14 we again see that our method for building dictionaries is much better than random selection.
The classification error rates for D from \(\hbox {D}_{0.28\%}\) to \(\hbox {D}_{5.82\%}\) for BIDMC Congestive Heart Failure Database (www.physionet.org/physiobank/database/chfdb/)
We again test the Uniform Scaling distance instead of Euclidean distance in both the training/testing phases. Based on studies of variability for human heartbeats (www.physionet.org/physiobank/database/chfdb/; http://en.wikipedia.org/wiki/Electrocardiography) and advice from a cardiologist, we chose a maximum scaling factor of 25 %. In Fig. 15, Uniform Scaling obtains a 0.035 testing error rate at \(\mathbf{D}_{{\varvec{4.6\%}}}\), significantly better than using the Euclidean distance.
As illustrated in Fig. 7, the threshold distance for D is 7.1. With this threshold, the algorithm rejects 4.8 % of the testing queries. Once again, these rejections (which can be seen at https://sites.google.com/site/dmkdrealistic/) all seem like reasonable rejections due to loss of signal or extraordinary amounts of noise/machine artifacts.
4.3 An example application in daily acitivies
We apply our framework to a widely studied benchmark dataset from MIT that contains 20 subjects performing approximately 30 h of daily activities (Bao and Intille 2004), such as running, stretching, scrubbing, vacuuming, riding-escalator, brushing-teeth, walking, bicycling, etc. The data was sampled at 70 Hz. We randomly chose 50 % of the data as training data, and treated the rest as testing data.
In Fig. 16, we show the training/testing error rates as our algorithm grows the data dictionary from the smallest size \((\mathbf{D}_{{\varvec{0.17\%}} })\) to the point where it is clear that our algorithm no longer improves. The use-all-the-training-data strawman (Bao and Intille 2004; Pham et al. 2010; Reiss and Stricker 2011), has a testing error rate of 0.237; however, we duplicate this error rate at \(\mathbf{D}_{{\varvec{1.1\%}}}\) and obtain the significantly lower error rate of 0.152 at \(\mathbf{D}_{{\varvec{3.8\%}}}\).
We also compare with the widely used rival approach discussed in Sect. 4.1 (Bao and Intille 2004; Reiss and Stricker 2011). The best result is error rate of 0.314 achieved by C4.5 decision tree (https://sites.google.com/site/dmkdrealistic/).
The classification error rates for D from \(\hbox {D}_{0.17\%}\) to \(\hbox {D}_{5.32\%}\) for the  Bao and Intille (2004)
Bao and Intille (2004)
In Fig. 17, we show that using Uniform Scaling distance again beats Euclidean distance, obtaining a mere 0.091 testing error rate at \(\mathbf{D}_{{\varvec{4.6\%}}}\). The threshold learned for D is 13.5, which rejects 6.3 % of the testing queries (https://sites.google.com/site/dmkdrealistic/).
The  curves are train/test error rates obtained when we replaced Euclidean distance with Uniform Scaling distance. Note the other curves are taken from Fig. 16 for comparison purposes (Color figure online)
curves are train/test error rates obtained when we replaced Euclidean distance with Uniform Scaling distance. Note the other curves are taken from Fig. 16 for comparison purposes (Color figure online)
4.4 Improving the anytime properties using complexity as search order
To evaluate the performance of our proposed ordering method, we simulate the classification of queries with varying arrival rates k. For the purpose of generality over all datasets, the arrival rates are modeled in Eq. (3) as a function of the number of all the subsequences \(\mathtt{{\vert }subs{\vert }}\) in the data dictionary (Shieh and Keogh 2010). This is because using the concrete numerical values (e.g. the frequency of the data generated at 250 Hz) may not always be meaningful or applicable, due to the wide variability in dataset characteristics.
For k = 1, the arrival rate of the streaming queries is exactly the time needed to calculate all the data dictionary, which is the same amount of time for the sequential search in Table 1. For k = 0.1, the arrival time of the streaming queries is only one-tenth the time of calculating the entire data dictionary.
We show the classification results using our ordering approach and the round robin approach (Shieh and Keogh 2010) in Fig. 18. With the round robin approach, the classification accuracies decrease a lot with faster query arrival time. This result is generally expected and can be attributed to the reduced computation time on each query for faster streams. In comparison, the classification accuracy of our ordering approach does not decrease significantly with faster arrival time.
4.5 Time and space complexity analysis
The time complexity for training and testing is \(\hbox {O}\,(\hbox {N}^{2})\) and O (N), respectively. The reasoning is as follows:
In the training phase, we first equally split the training data into two sections, part A and B. From part A, we randomly sample a large number of queries with length w. In order to cover more training data, the number of queries is proportional to the size of the training data. We use the randomly sampled queries to rank each object in B. Using the algorithm in Table 3, we need to find the nearest neighbor friend and nearest neighbor enemy for each query. Thus, the running time it takes for ranking every point in B is \(\hbox {O}\,(\hbox {N}/2)*\hbox {O}\,(\hbox {N}/2)\sim \, \hbox {O}\,(\hbox {N}^{2})\). In addition, it will take a constant time to extract and put the subsequences with highest scores into the dictionary. Thus, the time complexity in the training phase is O \((\hbox {N}^{2})\).
We note that the training times are not that important in this case. Consider the two main datasets we considered in our experiments:
-
Physiology It took days to collect this data, minutes to train on it.
-
Cardiology It took 150 minutes (of sensor time) to collect this data, a few tens of minutes to train on it.
Thus, we can typically train on data faster than we can collect it. In fact, we could have made our training faster with various lower bounding, caching and indexing tricks (Keogh et al. 2004b), but having past the real-time threshold there is little motivation to do so.
In the testing phase, we perform a linear search for the nearest neighbors of the incoming queries on the data dictionary that was built in the training phase. The running time in the testing phase is O(N), although the size of the data dictionary is only a very small percent of the training data. As with the training phase, we could make at least constant time improvements to this using with various lower bounding, and indexing mechanisms (Keogh et al. 2004b), however we are already more than fast enough to classify gestures on a smartphone, while using only a tiny fraction of its computational resources (memory and CPU), thus we have little incentive to further optimize this.
The space complexity for both training and testing phases is O (N). The reason is that during training we need to keep record of the scores for every point in the training data. In the testing phase, we need a small space to store the dictionary, which is a very small fraction of the training data.
5 Related work
There is significant literature on time series classification (Bao and Intille 2004; Chen et al. 2005; Gafurov et al. 2006; Morse and Patel 2007; Ratanamahatana and Keogh 2004; Song and Kim 2006) both in the data mining community and beyond. However, almost all of these works make the four assumptions we relaxed in this work, and are thus orthogonal to the contributions here. Our algorithm can be seen as building a data dictionary of primitives for the very long streaming/continuous time series (Raptis et al. 2011, 2008). Other works have also done this, such as Raptis et al. (2008), but they use significant amount of human effort to hand-edit the time series into patterns. In contrast, we build dictionaries automatically, with no human intervention.
In the following, we hint at the wide existence of the unrealistic assumptions in literature, a more exhaustive would require a survey paper.
Many research efforts assume a large number of perfectly aligned atomic patterns are available. Our proposed concepts of weakly-labeled data and the data dictionary do not require such well-processed patterns. However, some researchers either propose non-trivial algorithms to extract such patterns from the original raw data or simply assume a data generation process to produce such patterns exists. For example, Hanson et al. (2009) notes, “... it is desirable to identify the boundaries of single gait cycles, or steps, and process them individually...” However, the task of segmenting the data can be more difficult than classifying them. In Hanson et al. (2009), the authors acknowledge this, “ Finding gait cycle boundaries requires identification of landmark features in the waveforms that occur each cycle. Natural gait variation and differences between normal and pathological gait make this task non-trivial.”
Researchers often “clean” datasets before publicly release them (Hu et al. 2013). This is a noble idea, but one that perhaps shields the community from the realities of real-world deployment. Indeed, authors have been critiqued for releasing less than ideal data. For example, authors in Zhang and Sawchuk (2012) criticize the UC-Berkeley WARD dataset (Yang et al. 2009) by noting “part of the sensed data is missed due to battery failure”.
There are many examples of human intervention of the data generation procedure to produce the perfectly aligned data. For example, Zhang and Sawchuk (2012) has a very rigid data generation process, by noting that “When the subject was asked to perform a trial of one specific activity, an observer standing nearby marked the starting and ending points of the period of the activity performed.” In addition, the subject was asked to repeat each activity multiple times. However, in most real-world scenarios, we cannot expect people to perform daily activities in this way.
Another widely existing unrealistic assumption is that the patterns to be classified are all of equal length (Hanson et al. 2009; Keogh et al. 2006; Koch et al. 2010; Pärkkä et al. 2006; Reiss and Stricker 2011). The most famous and widely used time series benchmark is the UCR archive (Keogh et al. 2006). All the forty-seven datasets are well preprocessed and are of equal length. However, in reality, patterns can be of different lengths. For example, the human heart rate can be different. People can walk at different speeds, etc. The authors in Koch et al. (2010) observed, “It is clearly visible that despite the normalization steps taken, there is still considerable variation within the same gesture type from the same person.”
The assumption that exists in almost all the time series classification literature is that they assume every item to be classified belongs to exactly one of the well-defined classes (Gafurov and Snekkenes 2008; Keogh et al. 2006; Pärkkä et al. 2006; Ratanamahatana and Keogh 2004). We can consider a typical example to demonstrate the wide existence of this assumption. For example, in Pärkkä et al. (2006), authors report the classification result of seven daily activities, lie, row, bike, sit/stand, run, nordic-walk, walk. However, in reality, there are much more human activities than the mentioned above. For example, hand-shake, open-the-door, etc. If the query with a concept other than the seven listed concepts, their classifier will still mistakenly report some class label. In our proposed framework, we use a rejection threshold to prevent this problem.
Our work is superficially similar to the idea of time series shapelets (Ye and Keogh 2009), which do not make assumption 1, that some external agency (almost invariably a human) has annotated the beginning and ending the relevant patterns. However, we can list some of the key differences between shapelets and the proposed work:
-
Shapelets discover a single pattern to represent each class. In this work, we learn a dictionary to represent each class.
-
Shapelets assume that every pattern belongs to some class. Here we make no such assumption.
-
Shapelets assume that every pattern is the same length. This work does not make that assumption; we simply give upper and lower bounds on how long the patterns can be. Moreover, different classes can have different length templates in their dictionary.
-
Shapelets assume that a single template can represent every class. This work does not assume every class can be represented by a single template. In other words, the classes can be polymorphic, as shown in Fig. 1.
Shapelets do have an advantage over our technique in allowing a visual intuition as to what makes the classes distinct from each other.
Finally while data-editing schemes have been applied to time series before (Ueno et al. 2010; Xi et al. 2006), the authors always explicitly made assumption ‘1’ and ‘2’, and implicitly seem to have made assumption ‘3’, thus these works are solving an intrinsically simpler and narrower problem.
6 Conclusion and future work
We introduced a novel framework that requires only very weakly-labeled data and removes the unjustified assumptions made in virtually all time series classification research. We demonstrated over several large, real-world datasets that our method is significantly more accurate than several common strawman algorithms. Moreover, with less than one tenth of the original data kept in D, we are at least ten times faster at classification time. Moreover, even if this ten-fold speed-up is not enough to keep up with the worse-case arrival rate, our casting of our technique as an anytime algorithm means that we can handles busts of query requests that are orders of magnitude faster than the typical arrival rate with little degradation in accuracy.
Our algorithm has one major parameter, the length of queries. In our activity datasets, we simply used the original authors values (Bao and Intille 2004; www.pamap.org/demo.html), and for ECGs we used a cardiologist’s suggestion. By changing these suggested values, we empirically found that we are not sensitive to this parameter. Nevertheless in future work, we plan to learn it directly from the data. When using the uniform scaling distance (instead of Euclidean distance) we must provide additional parameters of the maximum and minimum scaling we will allow. However, once again we found that the settings here are not too critical.
Notes
Note that only some ECG classification systems do beat extraction then classification (Faezipour et al. 2010). Many researchers believe that robust beat extraction can be a harder problem than classification itself (cf. Figs. 1 and 2), and thus present every subsequence extracted by a sliding window for classification. This is the approach we consider in Sect. 4.2, as we assume bedside monitoring.
Note the fact that the two patterns are out of phase does not make them non-redundant, as at query time only queries half their length are used, and they are sliding across the entire length of the patterns. Details in Sect. 4.2.
Where tractability is an issue, we may sample a subset of the queries.
We defer the discussion on how to choose a query length to Sect. 6.
The reader may ask why not Dynamic Time Warping? Empirically, we tried it and it does not help. Moreover, we should not expect it to help this problem; https://sites.google.com/site/dmkdrealistic/.
We only show the rejected queries in the first case study. See Project (https://sites.google.com/site/dmkdrealistic/) for examples of rejected queries from the other case studies.
Experimental results show that the threshold distances for D built with Euclidean distance and Uniform Scaling distance are almost identical. Therefore, we only report one threshold distance.
References
Andino SLG et al (2000) Measuring the complexity of time series: an application to neurophysiological signals. Hum Brain Map 11(1):46–57
Aspelin K (2005) Establishing pedestrian walking speeds. Portland State University. www.usroads.com/journals/p/rej/9710/re971001.htm. Accessed 24 Aug 2009
Aziz W, Arif M (2006) Complexity analysis of stride interval time series by threshold dependent symbolic entropy. EJAP 98(1):30–40
Batista G, Keogh E, Mafra-Neto A, Rowton E (2011) Sensors and software to allow computational entomology, an emerging application of data mining. SIGKDD demo paper
Batista G, Wang X, Keogh E (2011) A complexity-invariant distance measure for time series. In: SDM
Bao L, Intille SS (2004) Acitivity recognition from user-annotated acceleration data. In: Proceedings of the 2nd international conference on pervasive computing, pp 1–17
Cavagna GA, Heglund NC, Taylor CR (1977) Mechanical work in terrestrial locomotion: two basic mechanisms for minimizing energy expenditure. J Physiol 233(5):R243–R261
Chen L, Özsu MT, Oria V (2005) Robust and fast similarity search for moving object trajectories. In: Proceedings of the ACM SIGMOD
CMU Graphics Lab Motion Capture Database. www.mocap.cs.cmu.edu/. Accessed 24 April 2012
de Chazal P, O’Dwyer M, Reilly RB (2004) Automatic classification of ECG heartbeats using ECG morphology and heartbeat interval features. IEEE Trans Biomed Eng 51:1196-06
Electrocardiography, http://en.wikipedia.org/wiki/Electrocardiography
Faezipour M, Saeed A, Bulusu S, Nourani M, Minn H, Tamil L (2010) A patient-adaptive profiling scheme for ECG beat classification. IEEE Trans Inform Technol Biomed 14(5):1153–1165
Gafurov D, Helkala K, Søndrol T (2006) Biometric gait authentication using accelerometer sensor. J Comput 1(7):51–59
Gafurov D, Snekkenes E (2008) Towards understanding the uniqueness of gait biometric. In: 8th IEEE International Conference on Automatic Face & Gesture Recognition
Grass J, Zilberstein S (1995) Anytime algorithm development tools. Technical Report. UMI Order Number: UM-CS-1995-094, University of Massachusetts
Hanson MA, Powell Jr HC, Barth AT, Lach J, Brown MBC (2009) Neural network gait classification for on-body inerital sensors. In: Proceedings of the 2009 sixth international workshop on wearable and implantable body sensor networks
Hao Y, Chen Y, Zakaria J, Hu B, Rakthanmanon T, Keogh E (2013) Towards never-ending learning from time series streams. In: SIGKDD
Hu B, Chen Y, Keogh E (2013) Time series classification under more realistic assumptions. In: SDM
Hu B, Chen Y, Zakaria J, Ulanova L, Keogh E (2013) Classification of multi-dimensional streaming time series by weighting each classifier’s track record. In: ICDM
Hu B, Rakthanmanon TR, Hao Y, Evans S, Lonardi S, Keogh E (2011) Discovering the intrinsic cardinality and dimensionality of time series using MDL. In: ICDM
Keogh E, Zhu Q, Hu B, Hao Y, Xi X, Wei L, Ratanamahatana CA (2006) The UCR time series classification/clustering homepage. www.cs.ucr.edu/~eamonn/time_series_data/
Keogh E, Lonardi S, Ratanamahatana C (2004) Towards parameter-free data mining. In: Proceedings of the tenth ACM SIGKDD
Keogh E, Palpanas T, Zordan VB, Gunopulos D, Cardle M (2004) Indexing large human-motion databases. In: VLDB
Koch P, Konen W, Hein K (2010) Gesture recognition on few training data using slow feature analysis and parametric bootstrap. In: IJCNN
Kranen P, Seidl T (2009) Harnessing the strengths of anytime algorithms for constant data stremas. J Data Min Knowl Discov 19(2):245–260
Lester J, Choudhury T, Kern N, Borriello G, Hannaford B (2005) A hybrid discriminative/generative approach for modeling human activities. In: IJCAI
Li L, Prakash BA (2011) Time series clustering: complex is simpler. In: ICML
Li M, Vitanyi P (1997) An introduction to Kolmogorov complexity and its applications, 2nd edn. Springer Verlag, New York
Liu J, Yu K, Zhang Y, Huang Y (2010) Training conditional random fields using transfer learning for gesture recognition. In: ICDM
McMahon TA, Cheng GC (1990) The mechanics of running: how does stiffness couple with speed. J Biomech 23:65–78
Morse M, Patel JM (2007) An efficient and accurate method for evaluating time series similarity. In: Proceedings of SIGMOD
Niennattrakul V, Keogh E, Ratanamahatana CA (2010) Data editing techniques to allow the application of distance-based outlier detection to streams. In: ICDM
PAMAP, Physical activity monitoring for aging people. www.pamap.org/demo.html. Accessed 12 May 2012
Pärkkä J, Ermes M, Korpipää P, Mäntyjärvi J, Peltola J, Korhonen I (2006) Activity classification using realistic data from wearable sensors. IEEE Trans Inf Technol Biomed 10:119–128
Pekalska E, Duin RPW, Paclík P (2006) Prototype selection for dissimilarity-based classifiers. Pattern Recognit 39:189–208
Pham C, Plötz T, Olivier P (2010) A dynamic time warping approach to real-time activity recognition for food preparation. In: Proceedings of the first international joint conference on Ambient intelligence
Project URL: https://sites.google.com/site/dmkdrealistic/
Raptis M, Kirovski D, Hoppes H (2011) Real-time classification of dance gestures from skeleton animation. In: Proceedings of the ACM SIGGRAPH symposium on computer animation
Raptis M, Wnuk K, Soatto S (2008) Flexible dictionaries for action recognition. In: Proceedings of the 1st international workshop on machine learning for vision-based motion analysis
Rakthanmanon T, Keogh E, Lonardi S, Evans S (2011) Time series epenthesis: clustering time series streams requires ignoring some data. In: ICDM
Ratanamahatana CA (2012) Personal communcation. May 2012
Ratanamahatana CA, Keogh E (2004) Making time-series classification more accurate using learned constraints. In: SDM
Reiss A, Stricker D (2011) Introducing a modular activity monitoring system. In: 33th international EMBC
Shieh J, Keogh E (2010) Polishing the right apple: anytime classification also benefits data streams with constant arrival times. In: ICDM
Song J, Kim D (2006) Simultaneous gesture segmentation and recognition based on forward spotting accumulative HMM. In: Proceedings of the 18th ICPR
The BIDMC congestive heart failure database, www.physionet.org/physiobank/database/chfdb/
Ueno K, Xi X, Keogh E, Lee D (2010) Anytime classification using the nearest neighbor algorithm with applications to stream mining. In: ICDM
Usabiaga J, Bebis G, Erol A, Nicolescu M (2007) Recognizing simple human actions using 3D head movement. Comput Intell 23(4):484–496
Vatavu RD (2011) The effect of sampling rate on the performance of template-based gesture recognizers. In: Proceedings of ICMI
Xi X, Keogh E, Shelton C, Wei L, Ratanamahatana C (2006) Fast time series classification using numerosity reduction. In: ICML, pp 1033–1040
Ye L, Wang X, Keogh E, Mafra-Neto A (2009) Autocannibalistic and anyspace indexing algorithms with applications to sensor data mining. In: SDM
Ye L, Keogh E (2009) Time series shapelets: a new primitive for data mining. In: KDD, pp 947–956
Yang AY, Giani A, Giannatonio R, Gilani K et al (2009) Distributed human action recognition via wearable motion sensor networks. www.eecs.berkeley.edu/~yang/software/WAR/index.html
Yang K, Jiang H, Dong J, Zhang C, Wang Z (2012) An adaptive real-time method for fetal heart rate extraction based on phonocardiography. In: 2012 IEEE biomedical circuits and systems conference. BioCAS, pp 356–359
Zilberstein S, Russell S (1995) Approximate reasoning using anytime algorithms. In: Imprecise and approximate computation. Kluwer Academic Publishers, Dordrecht
Zhang M, Sawchuk AA (2012) USC-HAD: a daily activity recognition using wearable sensors. ACM international conference on ubiquitous computing (UbiComp) workshop on situation, activity and goal awareness(SAGAware)
Acknowledgments
We thank all the donors of datasets. We would like to acknowledge the financial support for our research provided by NSF Grants IIS – 1161997 and an award from Vodafone.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: G. Karypis.
Rights and permissions
About this article
Cite this article
Hu, B., Chen, Y. & Keogh, E. Classification of streaming time series under more realistic assumptions. Data Min Knowl Disc 30, 403–437 (2016). https://doi.org/10.1007/s10618-015-0415-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10618-015-0415-0




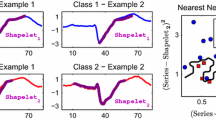
 , then there is little utility in adding any of the 14 or so other very similar patterns, including pattern
, then there is little utility in adding any of the 14 or so other very similar patterns, including pattern  . However, to robustly learn this concept (beats belonging to Record-08), we must add either example of the Premature Ventricular Contraction (PVC).
. However, to robustly learn this concept (beats belonging to Record-08), we must add either example of the Premature Ventricular Contraction (PVC).










 curves are train/test error rates obtained when we replaced Euclidean distance with Uniform Scaling distance (Color figure online)
curves are train/test error rates obtained when we replaced Euclidean distance with Uniform Scaling distance (Color figure online)



 curves are train/test error rates obtained when we replaced Euclidean distance with Uniform Scaling distance (Color figure online)
curves are train/test error rates obtained when we replaced Euclidean distance with Uniform Scaling distance (Color figure online)

