Abstract
Background
The spectrum of nonalcoholic fatty liver disease (NAFLD) describes disease conditions deteriorating from nonalcoholic fatty liver (NAFL) to nonalcoholic steatohepatitis (NASH) to cirrhosis (CIR) to hepatocellular carcinoma (HCC). From a molecular and biochemical perspective, our understanding of the etiology of this disease is limited by the broad spectrum of disease presentations, the lack of a thorough understanding of the factors contributing to disease susceptibility, and ethical concerns related to repeat sampling of the liver. To better understand the factors associated with disease progression, we investigated by next-generation RNA sequencing the altered expression of microRNAs (miRNAs) in liver biopsies of class III obese subjects (body mass index ≥40 kg/m2) biopsied at the time of elective bariatric surgery.
Methods
Clinical characteristics and unbiased RNA expression profiles for 233 miRs, 313 transfer RNAs (tRNAs), and 392 miscellaneous small RNAs (snoRNAs, snRNAs, rRNAs) were compared among 36 liver biopsy specimens stratified by disease severity.
Results
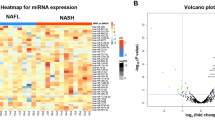
The abundances of 3 miRNAs that were found to be differentially regulated (miR-301a-3p and miR-34a-5p increased and miR-375 decreased) with disease progression were validated by RT-PCR. No tRNAs or miscellaneous RNAs were found to be associated with disease severity. Similar patterns of increased miR-301a and decreased miR-375 expression were observed in 134 hepatocellular carcinoma (HCC) samples deposited in The Cancer Genome Atlas (TCGA).
Conclusions
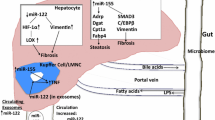
Our analytical results suggest that NAFLD severity is associated with a specific pattern of altered hepatic microRNA expression that may drive the hallmark of this disorder: altered lipid and carbohydrate metabolism. The three identified miRNAs can potentially be used as biomarkers to access the severity of NAFLD. The persistence of this miRNA expression pattern in an external validation cohort of HCC samples suggests that specific microRNA expression patterns may permit and/or sustain NAFLD development to HCC.

Similar content being viewed by others
References
Charlton M. Nonalcoholic fatty liver disease: a review of current understanding and future impact. Clin Gastroenterol Hepatol Off Clin Pract J Am Gastroenterol Assoc. 2004;2(12):1048–58.
Chalasani N, Younossi Z, Lavine JE, et al. The diagnosis and management of non-alcoholic fatty liver disease: practice guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology. 2012;55(6):2005–23.
Ekstedt M, Franzen LE, Mathiesen UL, et al. Long-term follow-up of patients with NAFLD and elevated liver enzymes. Hepatology. 2006;44(4):865–73.
Cheung O, Puri P, Eicken C, et al. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. 2008;48(6):1810–20.
Vickers KC, Roteta LA, Hucheson-Dilks H, Han L, Guo Y. Mining diverse small RNA species in the deep transcriptome. Trends Biochem Sci. 2015;40(1):4–7.
Han L, Vickers KC, Samuels DC, Guo Y. Alternative applications for distinct RNA sequencing strategies. Brief Bioinform. 2014;16(4):629–39.
Frye M, Watt FM. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr Biol. 2006;16(10):971–81.
Berg M, Agesen TH, Thiis-Evensen E, et al. Distinct high resolution genome profiles of early onset and late onset colorectal cancer integrated with gene expression data identify candidate susceptibility loci. Mol Cancer. 2010;9:100.
Bartlett JM, Thomas J, Ross DT, et al. Mammostrat as a tool to stratify breast cancer patients at risk of recurrence during endocrine therapy. Breast Cancer Res. 2010;12(4):R47.
Wei FY, Suzuki T, Watanabe S, et al. Deficit of tRNA(Lys) modification by Cdkal1 causes the development of type 2 diabetes in mice. J Clin Invest. 2011;121(9):3598–608.
Kirchhoff K, Machicao F, Haupt A, et al. Polymorphisms in the TCF7L2, CDKAL1 and SLC30A8 genes are associated with impaired proinsulin conversion. Diabetologia. 2008;51(4):597–601.
Omori S, Tanaka Y, Takahashi A, et al. Association of CDKAL1, IGF2BP2, CDKN2A/B, HHEX, SLC30A8, and KCNJ11 with susceptibility to type 2 diabetes in a Japanese population. Diabetes. 2008;57(3):791–5.
Yasukawa T, Suzuki T, Ueda T, Ohta S, Watanabe K. Modification defect at anticodon wobble nucleotide of mitochondrial tRNAs(Leu)(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes. J Biol Chem. 2000;275(6):4251–7.
Yasukawa T, Kirino Y, Ishii N, et al. Wobble modification deficiency in mutant tRNAs in patients with mitochondrial diseases. FEBS Lett. 2005;579(13):2948–52.
Kirino Y, Goto Y, Campos Y, Arenas J, Suzuki T. Specific correlation between the wobble modification deficiency in mutant tRNAs and the clinical features of a human mitochondrial disease. Proc Natl Acad Sci USA. 2005;102(20):7127–32.
Kleiner DE, Brunt EM, Van Natta M, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41(6):1313–21.
Fujii M, Shibazaki Y, Wakamatsu K, et al. A murine model for non-alcoholic steatohepatitis showing evidence of association between diabetes and hepatocellular carcinoma. Med Mol Morphol. 2013;46(3):141–52.
Guo Y, Bosompem A, Mohan S, et al. Transfer RNA detection by small RNA deep sequencing and disease association with myelodysplastic syndromes. BMC Genom. 2015;16:727.
Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. 2011;17:10–2.
Guo Y, Zhao S, Sheng Q, et al. Multi-perspective quality control of Illumina exome sequencing data using QC3. Genomics. 2014;103(5–6):322–8.
Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25.
Berezikov E, Robine N, Samsonova A, et al. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res. 2011;21(2):203–15.
Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Gene Dev. 2006;20(24):3407–25.
Westholm JO, Ladewig E, Okamura K, Robine N, Lai EC. Common and distinct patterns of terminal modifications to mirtrons and canonical microRNAs. RNA. 2012;18(2):177–92.
Larter CZ, Yeh MM. Animal models of NASH: getting both pathology and metabolic context right. J Gastroenterol Hepatol. 2008;23(11):1635–48.
Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37(Database issue):D93–7.
Flicek P, Amode MR, Barrell D, et al. Ensembl 2014. Nucleic Acids Res. 2014;42(Database issue):D749–55.
Guo Y, Zhao S, Ye F, Sheng Q, Shyr Y. MultiRankSeq: multiperspective approach for RNAseq differential expression analysis and quality control. Biomed Res Int. 2014;2014:248090.
Zou H, Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc B. 2005;67:301–20.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550.
Tibshirani R. Regression shrinkage and selection via the Lasso. J Roy Stat Soc B Met. 1996;58(1):267–88.
Domencich TA, McFadden D, Charles River Associates. Urban travel demand : a behavioral analysis : a Charles River Associates research study. New York: American Elsevier; 1975. vol xv, p. 215.
Hirsch RM, Slack JR, Smith RA. Techniques of trend analysis for monthly water-quality data. Water Resour Res. 1982;18(1):107–21.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8.
Wattacheril J, Seeley EH, Angel P, et al. Differential intrahepatic phospholipid zonation in simple steatosis and nonalcoholic steatohepatitis. PLoS One. 2013;8(2):e57165.
Guo Y, Bosompem A, Zhong X, Clark T, Shyr Y, Kim AS. A comparison of microRNA sequencing reproducibility and noise reduction using mirVana and TrIzol isolation methods. Int J Comput Biol Drug Des. 2014;7(2–3):102–12.
Odiase EO. The risk level of liver cancer cases among smokers in developing countries. Eur J Cancer. 2012;48:S13.
Varnholt H. The role of microRNAs in primary liver cancer. Ann Hepatol. 2008;7(2):104–13.
Jiang J, Gusev Y, Aderca I, et al. Association of microRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14(2):419–27.
Ladeiro Y, Couchy G, Balabaud C, et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47(6):1955–63.
Wong QWL, Lung RWM, Law PTY, et al. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology. 2008;135(1):257–69.
Li WQ, Chen C, Xu MD, et al. The rno-miR-34 family is upregulated and targets ACSL1 in dimethylnitrosamine-induced hepatic fibrosis in rats. FEBS J. 2011;278(9):1522–32.
Concepcion CP, Han YC, Mu P, et al. Intact p53-dependent responses in miR-34-deficient mice. PLoS Genet. 2012;8(7):e1002797.
Chang Y, Yan W, He XX, et al. miR-375 inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions. Gastroenterology. 2012;143(1):177-U357.
Liu AM, Poon RTP, Luk JM. MicroRNA-375 targets Hippo-signaling effector YAP in liver cancer and inhibits tumor properties. Biochem Bioph Res Co. 2010;394(3):623–7.
Ascha MS, Hanouneh IA, Lopez R, Tamimi TA, Feldstein AF, Zein NN. The incidence and risk factors of hepatocellular carcinoma in patients with nonalcoholic steatohepatitis. Hepatology. 2010;51(6):1972–8.
Takuma Y, Nouso K. Nonalcoholic steatohepatitis-associated hepatocellular carcinoma: our case series and literature review. World J Gastroenterol. 2010;16(12):1436–41.
Yasui K, Hashimoto E, Komorizono Y, et al. Characteristics of patients with nonalcoholic steatohepatitis who develop hepatocellular carcinoma. Clin Gastroenterol Hepatol. 2011;9(5):428–33 (e50).
Pirola CJ, Fernandez Gianotti T, Castano GO, et al. Circulating microRNA signature in non-alcoholic fatty liver disease: from serum non-coding RNAs to liver histology and disease pathogenesis. Gut. 2015;64(5):800–12.
Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One. 2011;6(8):e23937.
Yamada H, Suzuki K, Ichino N, et al. Associations between circulating microRNAs (miR-21, miR-34a, miR-122 and miR-451) and non-alcoholic fatty liver. Clin Chim Acta. 2013;424:99–103.
Tan Y, Ge G, Pan T, Wen D, Gan J. A pilot study of serum microRNAs panel as potential biomarkers for diagnosis of nonalcoholic fatty liver disease. PLoS One. 2014;9(8):e105192.
Celikbilek M, Baskol M, Taheri S, et al. Circulating microRNAs in patients with non-alcoholic fatty liver disease. World J Hepatol. 2014;6(8):613–20.
Estep M, Armistead D, Hossain N, et al. Differential expression of miRNAs in the visceral adipose tissue of patients with non-alcoholic fatty liver disease. Aliment Pharmacol Ther. 2010;32(3):487–97.
Sharma H, Estep M, Birerdinc A, et al. Expression of genes for microRNA-processing enzymes is altered in advanced non-alcoholic fatty liver disease. J Gastroenterol Hepatol. 2013;28(8):1410–5.
Jiang ZC, Tang XM, Zhao YR, Zheng L. A functional variant at miR-34a binding site in toll-like receptor 4 gene alters susceptibility to hepatocellular carcinoma in a Chinese Han population. Tumour Biol. 2014;35(12):12345–52.
Li W, Xie L, He X, et al. Diagnostic and prognostic implications of microRNAs in human hepatocellular carcinoma. Int J Cancer. 2008;123(7):1616–22.
Castro RE, Ferreira DM, Afonso MB, et al. miR-34a/SIRT1/p53 is suppressed by ursodeoxycholic acid in the rat liver and activated by disease severity in human non-alcoholic fatty liver disease. J Hepatol. 2013;58(1):119–25.
Min HK, Kapoor A, Fuchs M, et al. Increased hepatic synthesis and dysregulation of cholesterol metabolism is associated with the severity of nonalcoholic fatty liver disease. Cell Metab. 2012;15(5):665–74.
Streba LA, Vere CC, Rogoveanu I, Streba CT. Nonalcoholic fatty liver disease, metabolic risk factors, and hepatocellular carcinoma: an open question. World J Gastroenterol. 2015;21(14):4103–10.
Zhou P, Jiang W, Wu L, Chang R, Wu K, Wang Z. miR-301a is a candidate oncogene that targets the homeobox gene Gax in human hepatocellular carcinoma. Dig Dis Sci. 2012;57(5):1171–80.
Chang Y, Yan W, He X, et al. miR-375 inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions. Gastroenterology. 2012;143(1):177–87.
Li N, Fu H, Tie Y, et al. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 2009;275(1):44–53.
Yang P, Li QJ, Feng Y, et al. TGF-beta-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell. 2012;22(3):291–303.
He XX, Chang Y, Meng FY, et al. MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene. 2012;31(28):3357–69.
Liu AM, Poon RT, Luk JM. MicroRNA-375 targets Hippo-signaling effector YAP in liver cancer and inhibits tumor properties. Biochem Biophys Res Commun. 2010;394(3):623–7.
Robertson CL, Srivastava J, Siddiq A, et al. Astrocyte elevated gene-1 (AEG-1) regulates lipid homeostasis. J Biol Chem. 2015;290(29):18227–36.
Septer S, Edwards G, Gunewardena S, et al. Yes-associated protein is involved in proliferation and differentiation during postnatal liver development. Am J Physiol Gastrointest Liver Physiol. 2012;302(5):G493–503.
Acknowledgments
The National Institute of Diabetes And Digestive and Kidney Diseases of the National Institutes of Health supported the research reported in this publication, specifically through NIH grants DK020593 (Vanderbilt Diabetes Research and Training Center), 5UL1 RR024975-03 (CTSA), P30 CA68485 (Vanderbilt Ingram Cancer Center), DK058404 (Vanderbilt Digestive Disease Research Center), and R01 DK091748. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Guo, Y., Xiong, Y., Sheng, Q. et al. A micro-RNA expression signature for human NAFLD progression. J Gastroenterol 51, 1022–1030 (2016). https://doi.org/10.1007/s00535-016-1178-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00535-016-1178-0