Abstract
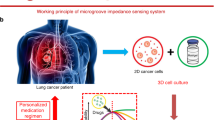
To predict the response of in vivo tumors, in vitro culture of cell colonies was suggested to be a standard assay to achieve high clinical relevance. To describe the responses of cell colonies, the most widely used quantification method is to count the number and size of cell colonies under microscope. That makes the colony formation assay infeasible to be high throughput and automated. In this work, in situ analysis of cell colonies suspended in soft hydrogel was developed based on impedance measurement technique. Cell colonies cultured between a pair of parallel plate electrodes were successfully analyzed by coating a layer of base hydrogel on one side of electrode. Real-time and label-free monitoring of cell colonies was realized during the culture course. Impedance magnitude and phase angle respectively represented the summation effect of colony responses and size of colonies. In addition, dynamic response of drug-treated colonies was demonstrated. High throughput and automatic colony formation assay was realized to facilitate more objective assessments in cancer research.

High throughput and automatic colony formation assay was realized by in situ impedimetric analysis across a pair of parallel plate electrodes in a culture chamber. Cell colonies suspended in soft hydrogel were cultured under the tested substance and their dynamic response was represented by impedance data.




Similar content being viewed by others
References
Abbott A. Cell culture: biology’s new dimension. Nature. 2003;424:870–2.
Pampaloni F, Reynaud EG, Stelzer EHK. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mole Cell Bio. 2007;8:839–45.
Weaver VM, Peterson OW, Wang F, Larabell CA, Briand P, Damsky C, et al. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137:312–8.
Santini MT, Rainaldi G. Three-dimensional spheroid model in tumor biology. Pathobiology. 1999;67:148–57.
Lin RZ, Chang HY. Recent advances in three-dimensional multicellular spheroid culture for biomedical research. Biotechnol J. 2008;3:1172–84.
Olive PL, Durand RE. Drug and radiation resistance in spheroids: cell contact and kinetics. Cancer Metast Rev. 1994;13:121.
Lee HJ, Zhuang G, Cao Y, Du P, Kim HJ, Settleman J. Drug resistance via feedback activation of stat3 in oncogene-addicted cancer cells. Cancer Cell. 2014;26:207–21.
Essig A, Duque-Afonso J, Schwemmers S, Pahl HL, Lübbert M. The AML1/ETO target gene LAT2 interferes with differentiation of normal hematopoietic precursor cells. Leukemia Res. 2014;38:340–5.
Jin L, Feng T, Shih HP, Zerda R, Luo A, Hsu J, et al. Colony-forming cells in the adult mouse pancreas are expandable in matrigel and form endocrine/acinar colonies in laminin hydrogel. Proc Natl Acad Sci U S A. 2013;110:3907–12.
Raghavan S, Ward MR, Rowley KR, Wold RM, Takayama S, Buckanovich RJ, et al. Formation of stable small cell number three-dimensional ovarian cancer spheroids using hanging drop arrays for preclinical drug sensitivity assays. Gynecol Oncol. 2015;138:181–9.
Lee IC, Chang JF. Label-free selection and enrichment of liver cancer stem cells by surface niches build up with polyelectrolyte multilayer films. Colloid Surf B. 2015;125:120–6.
Patra B, Peng CC, Liao WH, Lee CH, Tung YC. Drug testing and flow cytometry analysis on a large number of uniform sized tumor spheroids using a microfluidic device. Sci Rep. 2016;6:21061.
Stett A, Egert U, Guenther E, Hofmann F, Meyer T, Nisch W, et al. Biological application of microelectrode arrays in drug discovery and basic research. Anal Bioanal Chem. 2003;377:486–95.
Lei KF. Review on impedance detection of cellular responses in micro/nano environment. Micromachines. 2014;5:1–12.
Liu L, Xiao X, Lei KF, Huang CH. Quantitative impedimetric monitoring of cell migration under the stimulation of cytokine or anti-cancer drug in a microfluidic chip. Biomicrofluidics. 2015;9:034109.
Lei KF, Tseng HP, Lee CY, Tsang NM. Quantitative study of cell invasion process under extracellular stimulation of cytokine in a microfluidic device. Sci Rep. 2016;6:25557.
Giaever I, Keese CR. Monitoring fibroblast behavior in tissue culture with an applied electric field. Proc Natl Acad Sci U S A. 1984;81:3761–4.
Ehret R, Baumann W, Brischwein M, Schwinde A, Stegbauer K, Wolf B. Monitoring of cellular behavior by impedance measurements on interdigitated electrode structures. Biosens Bioelectron. 1997;12:29–41.
Hong J, Kandasamy K, Marimuthu M, Choi CS, Kim S. Electrical cell-substrate impedance sensing as a non-invasive tool for cancer cell study. Analyst. 2011;136:237–45.
Man I, Szebeni GJ, Plangar I, Szabo ER, Tokes T, Szabo Z, et al. Novel real-time cell analysis platform for the dynamic monitoring of ionizing radiation effects on human tumor cell lines and primary fibroblasts. Mol Med Rep. 2015;12:4610–9.
Dowling CM, Herranz Ors CH, Kiely PA. Using real-time impedance-based assays to monitor the effects of fibroblast-derived media on the adhesion, proliferation, migration and invasion of colon cancer cells. Biosci Rep. 2014;34:e00126.
Lei KF, Wu MH, Hsu CW, Chen YD. Real-time and non-invasive impedimetric monitoring of cell proliferation and chemosensitivity in a perfusion 3D cell culture microfluidic chip. Biosens Bioelectron. 2014;51:16–21.
Longati P, Jia X, Eimer J, Wagman A, Witt MR, Rehnmark S, et al. 3D pancreatic carcinoma spheroids induce a matrix-rich, chemoresistant phenotype offering a better model for drug testing. BMC Cancer. 2013;13:95.
Liu J, Tan Y, Zhang H, Zhang Y, Xu P, Chen J, et al. Soft fibrin gels promote selection and growth of tumorigenic cells. Nature Mater. 2012;11:734–41.
Lei KF, Wu ZM, Huang CH. Impedimetric quantification of the formation process and the chemosensitivity of cancer cell colonies suspended in 3D environment. Biosens Bioelectron. 2015;74:878–85.
Lee TKW, Lau TCM, Ng IOL. Doxorubicin-induced apoptosis and chemosensitivity in hepatoma cell lines. Cancer Chemother Pharmacol. 2002;49:78–86.
Chen MB, Wu XY, Gu JH, Guo QT, Shen WX, Lu PH. Activation of AMP-activated protein kinase contributes to doxorubicin-induced cell death and apoptosis in cultured myocardial H9c2 cells. Cell Biochem Biophys. 2011;60:311–22.
Sabatier R, GonçalvesAuthor Vitae A, Bertucci F. Personalized medicine: present and future of breast cancer management. Crit Rev Oncol Hemat. 2014;91:223–33.
Jackson SE, Chester JD. Personalised cancer medicine. Int J Cancer. 2015;137:262–6.
Kamiyama H, Rauenzahn S, Shim JS, Karikari CA, Feldmann G, Hua L, et al. Personalized chemotherapy profiling using cancer cell lines from selectable mice. Clin Cancer Res. 2015;19:1139–46.
Acknowledgements
This work was supported by Ministry of Science and Technology, Taiwan (Project no. MOST103-2221-E-182-004-MY3) and Chang Gung Memorial Hospital, Linkou Branch, Taiwan (Project no. CMRPG3C1921 and BMRPC05).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Lei, K.F., Kao, CH. & Tsang, NM. High throughput and automatic colony formation assay based on impedance measurement technique. Anal Bioanal Chem 409, 3271–3277 (2017). https://doi.org/10.1007/s00216-017-0270-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-017-0270-5