Abstract
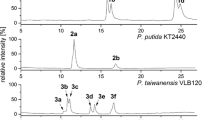
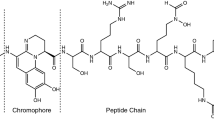
High-affinity iron (Fe)-scavenging molecules, or siderophores, are secreted by microorganisms to acquire and compete for Fe. Pyoverdine (PVD), the primary siderophore produced by Pseudomonas, consists of a dihydroxyquinoline-type chromophore, a peptide chain of variable length and conformation, and a side chain composed of a dicarboxylic acid or its monoamide derivative. Elucidation of the PVD structures secreted by different Pseudomonas strains is an important step toward understanding their Fe-transport strategies. In this study, we characterized multiple PVDs secreted by Pseudomonas putida KT2440 and Pseudomonas fluorescens RA12 using ultra-high performance liquid chromatography coupled with high-resolution quadrupole-orbitrap tandem mass spectrometry. To avoid purification steps prior to characterizing the bacterial supernatants, PVD candidates were identified by extracting fragments of the dihydroxyquinoline component from the chromatographic peaks. Varying collisional dissociation energies were subsequently applied to achieve, with high mass accuracy, a broad coverage of fragments of the entire PVD. Our approach allowed us to discriminate between three different PVD structures in the secretion of each strain. The three PVDs of P. putida possess the same peptide chain of seven amino acids, Asp-Orn-OHAsp-Dab-Gly-Ser-cOHOrn, with a cyclicized portion present in two of the PVDs. For P. fluorescens, two of the PVDs had the same peptide chain of 13 amino acids, Ala-Lys-Gly-Gly-Ala-OHAsp-Gly-Ser-Ala-Ala-Ala-Ala-cOHOrn, whereas a third PVD had a Ser substituting for the first Ala. The side chain of the PVDs was either succinic acid or succinamide. The present approach can be employed for simultaneous structural characterization of several peptidic siderophores and related molecules in bacterial secretions.

Characterizing mutiple pyoverdine (PVD) structures in bacterial secretions without prepurification step





Similar content being viewed by others
References
Poblete-Castro I, Becker J, Dohnt K, dos Santos V, Wittmann C (2012) Industrial biotechnology of Pseudomonas putida and related species. Appl Microbiol Biotechnol 93(6):2279–2290
Spiers AJ, Buckling A, Rainey PB (2000) The causes of Pseudomonas diversity. Microbiology 146(10):2345–2350
Silby MW, Winstanley C, Godfrey SAC, Levy SB, Jackson RW (2011) Pseudomonas genomes: diverse and adaptable. FEMS Microbiol Rev 35(4):652–680
Cezard C, Farvacques N, Sonnet P (2015) Chemistry and biology of Pyoverdines, pseudomonas primary siderophores. Curr Med Chem 22(2):165–186
Cornelis P, Matthijs S (2007) Pseudomonas siderophores and their biological significance. In: Varma A, Chincholkar S (eds) Microbial siderophores, vol 12. Soil biology. Springer, Berlin, pp 193–203
Meyer J-M (2000) Pyoverdines: pigments, siderophores and potential taxonomic markers of fluorescent Pseudomonas species. Arch Microbiol 174(3):135–142
Visca P, Imperi F, Lamont IL (2007) Pyoverdine siderophores: from biogenesis to biosignificance. Trends Microbiol 15(1):22–30
Cornelis P (2010) Iron uptake and metabolism in pseudomonads. Appl Microbiol Biotechnol 86(6):1637–1645
Matthijs S, Laus G, Meyer J-M, Abbaspour-Tehrani K, Schäfer M, Budzikiewicz H, Cornelis P (2009) Siderophore-mediated iron acquisition in the entomopathogenic bacterium Pseudomonas entomophila L48 and its close relative Pseudomonas putida KT2440. BioMetals 22(6):951–964
Meyer J-M, Gruffaz C, Raharinosy V, Bezverbnaya I, Schäfer M, Budzikiewicz H (2008) Siderotyping of fluorescent Pseudomonas: molecular mass determination by mass spectrometry as a powerful pyoverdine siderotyping method. BioMetals 21(3):259–271
Meyer J-M, Geoffroy VA, Baida N, Gardan L, Izard D, Lemanceau P, Achouak W, Palleroni NJ (2002) Siderophore typing, a powerful tool for the identification of fluorescent and nonfluorescent pseudomonads. Appl Environ Microbiol 68(6):2745–2753
Meyer J-M (2010) Pyoverdine siderophores as taxonomic and phylogenic markers. In: Ramos JL, Filloux A (eds) Pseudomonas. Springer, Netherlands, pp 201–233
Meyer J-M, Stintzi A, Coulanges V, Shivaji S, Voss JA, Taraz K, Budzikiewic H (1998) Siderotyping of fluorescent pseudomonads:characterization of pyoverdines of Pseudornonas fluorescens and Pseudornonas putida strains from Antarctica. Microbiology 144(11):3119–3126
Bultreys A, Gheysen I, Wathelet B, Maraite H, de Hoffmann E (2003) High-performance liquid chromatography analyses of pyoverdin siderophores differentiate among phytopathogenic fluorescent Pseudomonas species. Appl Environ Microbiol 69(2):1143–1153
Schäfer M, Fuchs R, Budzikiewicz H, Springer A, Meyer J-M, Linscheid M (2006) Structure elucidation of cyclic pyoverdins and examination of rearrangement reactions in MS/MS experiments by determination of exact product ion masses. J Mass Spectrom 41(9):1162–1170
Fuchs R, Budzikiewicz H (2001) Rearrangement reactions in the electrospray ionization mass spectra of pyoverdins. Int J Mass Spectrom 210–211:603–612
Fuchs R, Budzikiewicz H (2001) Structural studies of pyoverdins by mass spectrometry. Curr Org Chem 5(3):265–288
Budzikiewicz H, Schafer M, Fernandez DU, Meyer J (2006) Structure proposal for a new pyoverdin from Pseudomonas sp. PS 6.10. Z Naturforsch C 61(11/12):815
Domon B, Aebersold R (2006) Mass spectrometry and protein analysis. Science 312(5771):212–217
Aebersold R, Mann M (2003) Mass spectrometry-based proteomics. Nature 422(6928):198–207
Aebersold R, Goodlett DR (2001) Mass spectrometry in proteomics. Chem Rev 101(2):269–296
Eng J, McCormack A, Yates J (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5(11):976–989
Perkins DN, Pappin DJC, Creasy DM, Cottrell JS (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20(18):3551–3567
Steen H, Mann M (2004) The abc’s (and xyz’s) of peptide sequencing. Nat Rev Mol Cell Biol 5(9):699–711
Roepstorff P, Fohlman J (1984) Proposal for a nomenclature for sequence ions in mass spectra of peptides. Biomed Mass Spectrosc 11:601
Dongré AR, Jones JL, Somogyi Á, Wysocki VH (1996) Influence of peptide composition, gas-phase basicity, and chemical modification on fragmentation efficiency: evidence for the mobile proton model. J Am Chem Soc 118(35):8365–8374
Tang XJ, Thibault P, Boyd RK (1993) Fragmentation reactions of multiply-protonated peptides and implications for sequencing by tandem mass spectrometry with low-energy collision-induced dissociation. Anal Chem 65(20):2824–2834
Nilsson CL (2011) Advances in quantitative phosphoproteomics. Anal Chem 84(2):735–746
Jedrychowski MP, Huttlin EL, Haas W, Sowa ME, Rad R, Gygi SP (2011) Evaluation of HCD- and CID-type fragmentation within their respective detection platforms for murine phosphoproteomics. Mol Cell Proteomics 10(12):M111.009910
Frese CK, Altelaar AFM, Hennrich ML, Nolting D, Zeller M, Griep-Raming J, Heck AJR, Mohammed S (2011) Improved peptide identification by targeted fragmentation using CID, HCD and ETD on an LTQ-Orbitrap Velos. J Proteome Res 10(5):2377–2388
Shen Y, Tolić N, Xie F, Zhao R, Purvine SO, Schepmoes AA, Moore RJ, Anderson GA, Smith RD (2011) Effectiveness of CID, HCD, and ETD with FT MS/MS for degradomic-peptidomic analysis: comparison of peptide identification methods. J Proteome Res 10(9):3929–3943
Diedrich J, Pinto AM, Yates J III (2013) Energy dependence of HCD on peptide fragmentation: stepped collisional energy finds the sweet spot. J Am Soc Mass Spectrom 24(11):1690–1699
Neta P, Simon-Manso Y, Yang X, Stein SE (2009) Collisional energy dependence of peptide ion fragmentation. J Am Soc Mass Spectrom 20(3):469–476
Bushee JL, Argikar UA (2011) An experimental approach to enhance precursor ion fragmentation for metabolite identification studies: application of dual collision cells in an orbital trap. Rapid Commun Mass Spectrom 25(10):1356–1362
Scigelova M, Makarov A (2006) Orbitrap mass analyzer—overview and applications in proteomics. Proteomics 6(S2):16–21
Denisov E, Damoc E, Lange O, Makarov A (2012) Orbitrap mass spectrometry with resolving powers above 1,000,000. Int J Mass Spectrom 325–327:80–85
Zhang Y, Hao Z, Kellmann M, Huhmer A (2012) HR/AM targeted peptide quantitation on a Q Exactive MS: a unique combination of high selectivity, sensitivity and throughput. SRM 6:7
Lu W, Clasquin MF, Melamud E, Amador-Noguez D, Caudy AA, Rabinowitz JD (2010) Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone Orbitrap Mass Spectrometer. Anal Chem 82(8):3212–3221
Budzikiewicz H, Schäfer M, Fernández D, Matthijs S, Cornelis P (2007) Characterization of the chromophores of pyoverdins and related siderophores by electrospray tandem mass spectrometry. BioMetals 20(2):135–144
Hider RC, Kong X (2010) Chemistry and biology of siderophores. Nat Prod Rep 27(5):637–657
Ravel J, Cornelis P (2003) Genomics of pyoverdine-mediated iron uptake in pseudomonads. Trends Microbiol 11(5):195–200
Salah El Din ALM, Kyslík P, Stephan D, Abdallah MA (1997) Bacterial iron transport: structure elucidation by FAB-MS and by 2D NMR (1H, 13C, 15N) of pyoverdin G4R, a peptidic siderophore produced by a nitrogen-fixing strain of Pseudomonas putida. Tetrahedron 53(37):12539–12552
Mohn G, Taraz K, Budzikiewicz H, Naturforsch Z (1990) New pyoverdin-type siderophores from Pseudomonas fluorescens. Z Naturforsc B J Chem Sci 45(10):1437–1450
Acknowledgments
We are grateful to Dr. Rania Abou-Kandil for providing the P. fluorescens strain. We thank David F. Flannelly and Matthew A. Kukurugya for commenting on earlier versions of the manuscript. This work was supported in part by a Research Starter grant from the U.S. National Science Foundation (SSB 1337292) from the Division of Systems and Synthetic Biology.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 75 kb)
Rights and permissions
About this article
Cite this article
Wei, H., Aristilde, L. Structural characterization of multiple pyoverdines secreted by two Pseudomonas strains using liquid chromatography-high resolution tandem mass spectrometry with varying dissociation energies. Anal Bioanal Chem 407, 4629–4638 (2015). https://doi.org/10.1007/s00216-015-8659-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-015-8659-5