Abstract
Monitoring of intracellular redox status in a bacterial cell provides vital information about the physiological status of the cell, which can be exploited in several applications such as metabolic engineering and computational modeling. Fluorescent protein-based genetically encoded sensors can be used to monitor intracellular oxidation/reduction status. This study reports the development of a redox sensor for intracellular measurements using fluorescent protein pairs and the phenomenon of Förster resonance energy transfer (FRET). For the development of the sensor, fluorescent proteins Citrine and Cerulean were genetically modified to carry reactive cysteine residues on the protein surface close to the chromophore and a constructed FRET pair was fused using a biotinylation domain as a linker. In oxidized state, the FRET pairs are in close proximity by labile disulfide bond formation resulting in higher FRET efficiency. In reducing environment, the FRET is diminished due to the increased distance between FRET pairs providing large dynamic measurement range to the sensor. Intracellular studies in Escherichia coli mutants revealed the capability of the sensor in detecting real-time redox variations at single cell level. The results were validated by intensity based and time resolved measurements. The functional immobilization of the fluorescent protein-based FRET sensor at solid surfaces for in vitro applications was also demonstrated.

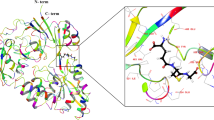
Schematic representation of FRET-based redox sensor







Similar content being viewed by others
References
Bob BB (1991) Regulation of CO2 assimilation in oxygenic photosynthesis: the ferredoxin/thioredoxin system: perspective on its discovery, present status, and future development. Arch Biochem Biophys 288:1
Rabinowitz JD, Vacchino JF, Beeson C, McConnell HM (1998) Potentiometric measurement of intracellular redox activity. J Am Chem Soc 120:2464
Barron JT, Gu L, Parrillo JE (2000) NADH/NAD redox state of cytoplasmic glycolytic compartments in vascular smooth muscle. Am J Physiol Heart Circ Physiol 279:H2872
Gorodetsky AA, Dietrich LEP, Lee PE, Demple B, Newman DK, Barton JK (2008) DNA binding shifts the redox potential of the transcription factor SoxR. Proc Natl Acad Sci U S A 105:3684
Ritz D, Beckwith J (2001) Roles of thiol-redox pathways in bacteria. Annu Rev Microbiol 55:21–48
Oktyabrsky ON, Smirnova GV (2007) Redox regulation of cellular functions. Biochemistry (Mosc) 72:132
Sen CK (2000) Cellular thiols and redox-regulated signal transduction. Curr Top Cell Regul 36:1
Brandes N, Schmitt S, Jakob U (2009) Thiol-based redox switches in eukaryotic proteins. Antioxid Redox Signal 11:997
Bykov VJ, Lambert JM, Hainaut P, Wiman KG (2009) Mutant p53 rescue and modulation of p53 redox state. Cell Cycle 8:2509
Yano T, Oku M, Akeyama N, Itoyama A, Yurimoto H, Kuge S, Fujiki Y, Sakai Y (2010) A novel fluorescent sensor protein for visualization of redox states in the cytoplasm and in peroxisomes. Mol Cell Biol 30:3758
Schafer FQ, Buettner GR (2001) Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med 30:1191
Kemp M, Go YM, Jones DP (2008) Nonequilibrium thermodynamics of thiol/disulfide redox systems: a perspective on redox systems biology. Free Radic Biol Med 44:921
Hwang C, Sinskey A, Lodish H (1992) Oxidized redox state of glutathione in the endoplasmic reticulum. Science 257:1496
Wardman P (2007) Fluorescent and luminescent probes for measurement of oxidative and nitrosative species in cells and tissues: progress, pitfalls, and prospects. Free Radic Biol Med 43:995
Tsien RY (1998) The green fluorescent protein. Annu Rev Biochem 67:509–544
Zhang J, Campbell RE, Ting AY, Tsien RY (2002) Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol 3:906
Shaner NC, Steinbach PA, Tsien RY (2005) A guide to choosing fluorescent proteins. Nat Methods 2:905
Ostergaard H, Henriksen A, Hansen FG, Winther JR (2001) Shedding light on disulfide bond formation: engineering a redox switch in green fluorescent protein. EMBO J 20:5853
Grynkiewicz G, Poenie M, Tsien RY (1985) A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem 260:3440
Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, Remington SJ (2004) Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J Biol Chem 279:13044
Hoi H, Ding Y, Campbell RE (2013) In: Medintz IL, Hildebrandt N (eds) FRET—Förster resonance energy transfer: from theory to applications. Wiley, Weinheim
Truong K, Sawano A, Miyawaki A, Ikura M (2007) Calcium indicators based on calmodulin-fluorescent protein fusions. Methods Mol Biol 352:71
VanEngelenburg SB, Palmer AE (2008) Fluorescent biosensors of protein function. Curr Opin Chem Biol 12:60
Miyawaki A (2011) Development of probes for cellular functions using fluorescent proteins and fluorescence resonance energy transfer. Annu Rev Biochem 80:357
Kolossov VL, Spring BQ, Sokolowski A, Conour JE, Clegg RM, Kenis PJA, Gaskins HR (2008) Engineering redox-sensitive linkers for genetically encoded FRET-based biosensors. Exp Biol Med 233:238
Reddy DV, Shenoy BC, Carey PR, Sonnichsen FD (2000) High resolution solution structure of the 1.3S subunit of transcarboxylase from Propionibacterium shermanii. Biochemistry 39:2509
Maniatis T, Fritsch EF, Sambrook J (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Griesbeck O, Baird GS, Campbell RE, Zacharias DA, Tsien RY (2001) Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J Biol Chem 276:29188
Rizzo MA, Springer GH, Granada B, Piston DW (2004) An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol 22:445
Santala V, Lamminmäki U (2004) Production of a biotinylated single-chain antibody fragment in the cytoplasm of Escherichia coli. J Immunol Methods 284:165
Abraham BG, Tkachenko NV, Santala V, Lemmetyinen H, Karp M (2011) Bidirectional fluorescence resonance energy transfer (FRET) in Mutated and Chemically Modified Yellow Fluorescent Protein (YFP). Bioconjug Chem 22:227
Santala S, Efimova E, Kivinen V, Larjo A, Aho T, Karp M, Santala V (2011) Improved triacylglycerol production in acinetobacter baylyi ADP1 by metabolic engineering. Microb Cell Factories 10:36
Clegg RM (1992) Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol 211:353
Wallrabe H, Periasamy A (2005) Imaging protein molecules using FRET and FLIM microscopy. Curr Opin Biotechnol 16:19
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671
Gavet O, Pines J (2010) Activation of cyclin B1–Cdk1 synchronizes events in the nucleus and the cytoplasm at mitosis. J Cell Biol 189:247
Suzuki M, Ito Y, Savage HE, Husimi Y, Douglas KT (2003) Intramolecular fluorescent resonance energy transfer (FRET) by BODIPY chemical modification of cysteine-engineered mutants of green fluorescent protein. Chem Lett 32:306
Liu J, Lu Y (2002) FRET study of a trifluorophore-labeled DNAzyme. J Am Chem Soc 124:15208
Gohlke C, Murchie AI, Lilley DM, Clegg RM (1994) Kinking of DNA and RNA helices by bulged nucleotides observed by fluorescence resonance energy transfer. Proc Natl Acad Sci U S A 91:11660
Clegg RM, Murchie AI, Zechel A, Lilley DM (1993) Observing the helical geometry of double-stranded DNA in solution by fluorescence resonance energy transfer. Proc Natl Acad Sci U S A 90:2994
Kolossov VL, Spring BQ, Clegg RM, Henry JJ, Sokolowski A, Kenis PJA, Gaskins HR (2011) Development of a high-dynamic range, GFP-based FRET probe sensitive to oxidative microenvironments. Exp Biol Med 236:681
Lam AJ, St-Pierre F, Gong Y, Marshall JD, Cranfill PJ, Baird MA, McKeown MR, Wiedenmann J, Davidson MW, Schnitzer MJ, Tsien RY, Lin MZ (2012) Improving FRET dynamic range with bright green and red fluorescent proteins. Nat Methods 9:1005–1012
Evers TH, Van Dongen EMWM, Faesen AC, Meijer EW, Merkx M (2006) Quantitative understanding of the energy transfer between fluorescent proteins connected via flexible peptide linkers. Biochemistry (N Y) 45:13183
Evers TH, Appelhof MAM, de Graaf-Heuvelmans PTHM, Meijer EW, Merkx M (2007) Ratiometric detection of Zn(II) using chelating fluorescent protein chimeras. J Mol Biol 374:411
Green J, Paget MS (2004) Bacterial redox sensors. Nat Rev Microbiol 2:954
Sarkar P, Koushik SV, Vogel SS, Gryczynski I, Gryczynski Z (2009) Photophysical properties of Cerulean and Venus fluorescent proteins. J Biomed Opt 14:034047
Patterson GH, Knobel SM, Sharif WD, Kain SR, Piston DW (1997) Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys J 73:2782
Campbell TN, Choy FYM (2001) The effect of pH on green fluorescent protein: a brief review. Mol Biol Today 2:1
Saeed IA, Ashraf SS (2009) Denaturation studies reveal significant differences between GFP and blue fluorescent protein. Int J Biol Macromol 45:236
Veselov AA, Abraham BG, Lemmetyinen H, Karp MT, Tkachenko NV (2012) Photochemical properties and sensor applications of modified yellow fluorescent protein (YFP) covalently attached to the surfaces of etched optical fibers (EOFs). Anal Bioanal Chem 402:1149
Elsliger M, Wachter RM, Hanson GT, Kallio K, Remington SJ (1999) Structural and spectral response of green fluorescent protein variants to changes in pH. Biochemistry (N Y) 38:5296
Saito K, Welker E, Scheraga HA (2001) Folding of a disulfide-bonded protein species with free thiol(s): competition between conformational folding and disulfide reshuffling in an intermediate of bovine pancreatic ribonuclease A. Biochemistry 40:15002
Olsen KN, Budde BB, Siegumfeldt H, Rechinger KB, Jakobsen M, Ingmer H (2002) Noninvasive measurement of bacterial intracellular pH on a single-cell level with green fluorescent protein and fluorescence ratio imaging microscopy. Appl Environ Microbiol 68:4145
Booth IR (1985) Regulation of cytoplasmic pH in bacteria. Microbiol Rev 49:359
Mamathambika BS, Bardwell JC (2008) Disulfide-linked protein folding pathways. Annu Rev Cell Dev Biol 24:211
Beckwith J (2007) What lies beyond Uranus? Preconceptions, ignorance, serendipity and suppressors in the search for biology’s secrets. Genetics 176:733
Hatahet F, Nguyen VD, Salo K, Ruddock L (2010) Disruption of reducing pathways is not essential for efficient disulfide bond formation in the cytoplasm of E. coli. Microb Cell Factories 9:67
Markwardt ML, Kremers G, Kraft CA, Ray K, Cranfill PJC, Wilson KA, Day RN, Wachter RM, Davidson MW, Rizzo MA (2011) An improved Cerulean fluorescent protein with enhanced brightness and reduced reversible photoswitching. PLoS ONE 6:e17896
Derman AI, Prinz WA, Belin D, Beckwith J (1993) Mutations that allow disulfide bond formation in the cytoplasm of Escherichia coli. Science 262:1744
Xiong S, Wang YF, Ren XR, Li B, Zhang MY, Luo Y, Zhang L, Xie QL, Su KY (2005) Solubility of disulfide-bonded proteins in the cytoplasm of Escherichia coli and its "oxidizing" mutant. World J Gastroenterol 11:1077
Bilitewski U (2006) Protein-sensing assay formats and devices. Anal Chim Acta 568:232
Wong LS, Khan F, Micklefield J (2009) Selective covalent protein immobilization: strategies and applications. Chem Rev 109:4025
Kwon Y, Coleman MA, Camarero JA (2006) Selective immobilization of proteins onto solid supports through split-intein-mediated protein trans-splicing. Angew Chem Int Ed 45:1726
Kufer SK, Dietz H, Albrecht C, Blank K, Kardinal A, Rief M, Gaub HE (2005) Covalent immobilization of recombinant fusion proteins with hAGT for single molecule force spectroscopy. Eur Biophys J 35:72
Yin J, Liu F, Li X, Walsh CT (2004) Labeling proteins with small molecules by site-specific posttranslational modification. J Am Chem Soc 126:7754
Acknowledgments
This work was supported by LASKEMO National Graduate Programme, Finland.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 276 kb)
Rights and permissions
About this article
Cite this article
Abraham, B.G., Santala, V., Tkachenko, N.V. et al. Fluorescent protein-based FRET sensor for intracellular monitoring of redox status in bacteria at single cell level. Anal Bioanal Chem 406, 7195–7204 (2014). https://doi.org/10.1007/s00216-014-8165-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-014-8165-1