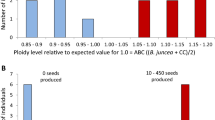
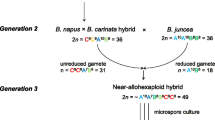
Abstract
Key message
Allohexaploid Brassica populations reveal ongoing segregation for fertility, while genotype influences fertility and meiotic stability.
Abstract
Creation of a new Brassica allohexaploid species is of interest for the development of a crop type with increased heterosis and adaptability. At present, no naturally occurring, meiotically stable Brassica allohexaploid exists, with little data available on chromosome behaviour and meiotic control in allohexaploid germplasm. In this study, 100 plants from the cross B. carinata × B. rapa (A2 allohexaploid population) and 69 plants from the cross (B. napus × B. carinata) × B. juncea (H2 allohexaploid population) were assessed for fertility and meiotic behaviour. Estimated pollen viability, self-pollinated seed set, number of seeds on the main shoot, number of pods on the main shoot, seeds per ten pods and plant height were measured for both the A2 and H2 populations and for a set of reference control cultivars. The H2 population had high segregation for pollen viability and meiotic stability, while the A2 population was characterised by low pollen fertility and a high level of chromosome loss. Both populations were taller, but had lower average fertility trait values than the control cultivar samples. The study also characterises fertility and meiotic chromosome behaviour in genotypes and progeny sets in heterozygous allotetraploid Brassica derived lines, and indicates that genotypes of the parents and H1 hybrids are affecting chromosome pairing and fertility phenotypes in the H2 population. The identification and characterisation of factors influencing stability in novel allohexaploid Brassica populations will assist in the development of this as a new crop species for food and agricultural benefit.









Similar content being viewed by others
References
Arumugam N, Mukhopadhyay A, Gupta V, Pental D, Pradhan AK (1996) Synthesis of hexaploid (AABBCC) somatic hybrids: a bridging material for transfer of ‘tour’ cytoplasmic male sterility to different Brassica species. Theor Appl Genet 92:762–768
Chen S, Nelson MN, Chèvre AM, Jenczewski E, Li Z, Mason AS et al (2011) Trigenomic bridges for Brassica improvement. Crit Rev Plant Sci 30:524–547
Cifuentes M, Eber F, Lucas MO, Lode M, Chèvre AM, Jenczewski E (2010a) Repeated polyploidy drove different levels of crossover suppression between homoeologous chromosomes in Brassica napus allohaploids. Plant Cell 22:2265–2276
Cifuentes M, Grandont L, Moore G, Chèvre AM, Jenczewski E (2010b) Genetic regulation of meiosis in polyploid species: new insights into an old question. New Phytol 186:29–36
Gaeta RT, Chris Pires J (2010) Homoeologous recombination in allopolyploids: the polyploid ratchet. New Phytol 186:18–28
Gaeta RT, Pires JC, Iniguez-Luy F, Leon E, Osborn TC (2007) Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19:3403–3417
Ge X-H, Wang J, Li Z-Y (2009) Different genome-specific chromosome stabilities in synthetic Brassica allohexaploids revealed by wide crosses with Orychophragmus. Ann Bot 104:19–31
Howard HW (1942) The effect of polyploidy and hybridity on seed size in crosses between Brassica chinensis, B. carinata, amphidiploid B. chinensis-carinata and autotetraploid B. chinensis. J Genet 43:105–119
Iwasa (1964) Cytogenetic studies on the artificially raised trigenomic hexaploid hybrid forms in the genus Brassica. J Fac Agric Kyushu Univ 13:309–352
Leitch AR, Letch IJ (2008) Genomic plasticity and the diversity of polyploid plants. Science 320:481–483
Li M, Qian W, Meng J, Li Z (2004) Construction of novel Brassica napus genotypes through chromosomal substitution and elimination using interploid species hybridisation. Chrom Res 12:417–426
Mason AS, Huteau V, Eber F, Coriton O, Yan G et al (2010) Genome structure affects the rate of autosyndesis and allosyndesis in AABC, BBAC and CCAB Brassica interspecific hybrids. Chrom Res 18:655–666
Mason AS, Nelson MN, Castello MC, Yan G, Cowling WA (2011) Genotypic effects on the frequency of homoeologous and homologous recombination in Brassica napus × B. carinata hybrids. Theor Appl Genet 122:543–553
Mason AS, Yan G, Cowling WA, Nelson MN (2012) A new method for producing allohexaploid Brassica through unreduced gametes. Euphytica 186:277–287
Mason AS, Batley J, Bayer PE, Hayward A, Cowling WA, Nelson MN (2014a) High-resolution molecular karyotyping uncovers pairing between ancestrally related Brassica chromosomes. New Phytol 202:964–974
Mason AS, Nelson MN, Takahira J, Cowling WA, Alves GM et al (2014b) The fate of chromosomes and alleles in an allohexaploid Brassica population. Genetics 197:273–283
Mason AS, Rousseau-Gueutin M, Morice J, Bayer PE, Besharat N et al (2016) Centromere locations in Brassica A and C genomes revealed through half-tetrad analysis. Genetics 202:513-523
Meng J, Shi S, Gan L, Li Z, Qu X (1998) The production of yellow-seeded Brassica napus (AACC) through crossing interspecific hybrids of B. campestris (AA) and B. carinata (BBCC) with B. napus. Euphytica 103:329–333
Morinaga T (1934) Interspecific hybridisation in Brassica. Cytologia 6:62–67
Nicolas S, Le Mignon G, Eber F, Coriton O, Monod H et al (2007) Homeologous recombination plays a major role in chromosome rearrangements that occur during meiosis of Brassica napus haploids. Genetics 175:487–503
Nicolas SD, Monod H, Eber F, Chèvre AM, Jenczewski E (2012) Non-random distribution of extensive chromosome rearrangements in Brassica napus depends on genome organization. Plant J 70:691–703
Pradhan A, Plummer JA, Nelson MN, Cowling WA, Yan G (2010) Successful induction of trigenomic hexaploid Brassica from a triploid hybrid of B. napus L. and B. nigra (L.) Koch. Euphytica 176:87–98
R Core Team (2015) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org
Rakow G (2004) Species origin and economic importance of Brassica. In: Pua EC, Douglas CJ (eds) Biotechnology in agriculture and forestry, vol 54. Springer, Berlin, pp 3–11
Ramsey J, Schemske DW (1998) Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu Rev Eco Syst 29:467–501
Sheidai M, Noormohamadi Z, Sotodeh M (2006) Cytogenetic variability in several canola (Brassica napus) cultivars. Caryologia 59:267–276
Sjödin C, Glimelius K (1989) Brassica naponigra, a somatic hybrid resistant to Phoma lingam. Theor Appl Genet 77:651–656
Song K, Lu PI, Tang K, Osborn TC (1995) Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc Natl Acad Sci USA 92:7719–7723
Song C, Liu S, Xiao J, He W, Zhou Y, Qin Q, Zhang C, Liu Y (2012) Polyploid organisms. Sci China Life Sci 1 55(4):301–311.
Szadkowski E, Eber F, Huteau V, Lodé M, Huneau C, Belcram H, Coriton O, Manzanares - Dauleux MJ, Delourme R, King GJ, Chalhoub B, Jenczewski E, Chèvre AM (2010) The first meiosis of resynthesized Brassica napus, a genome blender. New Phytol 186:102–112
Tian E, Jiang Y, Chen L, Zou J, Liu F, Meng J (2010) Synthesis of a Brassica trigenomic allohexaploid (B. carinata × B. rapa) de novo and its stability in subsequent generations. Theor Appl Genet 121:1431–1440
Udall JA, Wendell JF (2006) Polyploidy and crop improvement. Crop Sci 46(Supplement_1):S-3
UN (1935) Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilisation. Jap J Bot 7:389–452
Zhang G, Zhou W (2006) Genetic analyses of agronomic and seed quality traits of synthetic oilseed Brassica napus produced from interspecific hybridization of B. campestris and B. oleracea. J Genet 85:45–51
Zhou J, Tan C, Cui C, Ge X, Li Z (2016) Distinct subgenome stabilities in synthesized Brassica allohexaploids. Theor Appl Genet 129:1257–1271
Acknowledgements
This project, including a three month research visit by Margaret Mwathi from The University of Queensland, Brisbane, Australia to Punjab Agricultural University, Ludhiana, India was supported by an Australia–India Strategic Research Fund: Biotechnology grant (Project BF06520), jointly funded by the Australian Government Department of Industry, Innovation and Science and the Indian Government Department of Biotechnology. ASM is funded by Emmy Noether DFG Grant MA6473/1-1.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The authors declare that the study complies with the current laws of the country (India) in which they were performed.
Additional information
Communicated by Richard G. F. Visser.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2016_2850_MOESM1_ESM.pdf
Phenotypic traits (A, B, C and D) in an allohexaploid population (A2) derived from crosses between B. rapa and B. carinata compared against control cultivar samples B. carinata (PC5) and B. rapa (TL-17) (PDF 39 KB)
122_2016_2850_MOESM2_ESM.pdf
Phenotypic traits (A, B, C and D) in an allohexaploid population (H2) derived from the cross (B. napus × B. carinata) × B. juncea compared against control cultivars B. carinata (PC5), B. juncea (RLC-1) and B. napus (GSC-5) (PDF 38 KB)
122_2016_2850_MOESM3_ESM.pdf
Scatter plot matrix showing correlation and distributions of phenotypic traits between A. An allohexaploid population (A2) derived from crosses between B. rapa and B. carinata and B. An allohexaploid population (H2) derived from the cross (B. napus × B. carinata) × B. juncea (PDF 56 KB)
122_2016_2850_MOESM4_ESM.pdf
Genotypes in the H2 allohexaploid population derived from the cross (B. napus × B. carinata) × B. juncea, showing significant differences between genotypes in plant height, (p < 0.05, a and b Tukey’s HSD) (PDF 13 KB)
122_2016_2850_MOESM5_ESM.pdf
Progeny sets in the H2 allohexaploid population derived from the cross (B. napus × B. carinata) × B. juncea, showing significant differences in plant height,(p < 0.05, between progeny sets in “G1” and “G2” genotypes, between progeny sets in “G2”and “G3” genotype, and between progeny sets in “G1” and “G3” genotypes, Tukey’s HSD) (PDF 25 KB)
Rights and permissions
About this article
Cite this article
Mwathi, M.W., Gupta, M., Atri, C. et al. Segregation for fertility and meiotic stability in novel Brassica allohexaploids. Theor Appl Genet 130, 767–776 (2017). https://doi.org/10.1007/s00122-016-2850-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-016-2850-8