Abstract
Background
Diagnosis of bacterial meningitis remains a challenge in most developing countries due to low yield from bacterial culture, widespread use of non-prescription antibiotics, and weak microbiology laboratories. The objective of this study was to compare the yield from standard bacterial culture with the multiplex nested PCR platform, the BioFire® FilmArray® Meningitis/Encephalitis Panel (BioFire ME Panel), for cases with suspected acute bacterial meningitis.
Methods
Following Gram stain and bacterial culture on cerebrospinal fluid (CSF) collected from children aged less than 5 years with a clinical suspicion of acute bacterial meningitis (ABM) as defined by the WHO guidelines, residual CSF specimens were frozen and later tested by BioFire ME Panel.
Results
A total of 400 samples were analyzed. Thirty-two [32/400 (8%)] of the specimens were culture positive, consisting of; three Salmonella spp. (2 Typhi and 1 non-typhi), three alpha hemolytic Streptococcus, one Staphylococcus aureus, six Neisseria meningitidis, seven Hemophilus influenzae, 11 Streptococcus pneumoniae and 368 were culture negative. Of the 368 culture-negative specimens, the BioFire ME Panel detected at least one bacterial pathogen in 90 (24.5%) samples, consisting of S. pneumoniae, N. meningitidis and H. influenzae, predominantly. All culture positive specimens for H. influenzae, N. meningitidis and S. pneumoniae also tested positive with the BioFire ME Panel. In addition, 12 specimens had mixed bacterial pathogens identified. For the first time in this setting, we have data on the viral agents associated with meningitis. Single viral agents were detected in 11 (2.8%) samples while co-detections with bacterial agents or other viruses occurred in 23 (5.8%) of the samples.
Conclusions
The BioFire® ME Panel was more sensitive and rapid than culture for detecting bacterial pathogens in CSF. The BioFire® ME Panel also provided for the first time, the diagnosis of viral etiologic agents that are associated with meningoencephalitis in this setting. Institution of PCR diagnostics is recommended as a routine test for suspected cases of ABM to enhance early diagnosis and optimal treatment.
Similar content being viewed by others
Introduction
Acute bacterial meningitis (ABM) is the most common infection of the central nervous system and remains a major cause of mortality and morbidity in several countries in sub-Saharan Africa [1,2,3,4,5]. The annual estimate for the burden of ABM is 1.2 million each year worldwide [6,7,8]. While the incidence in most developed countries have declined significantly due to the wide implementation of pneumococcal and Haemophilus influenzae conjugate vaccines, such decline is yet to be accomplished in most developing countries where the bacterial etiologic agents are more diverse and diagnostic services are sparse. Further, the sporadic and unpredictable outbreaks of meningococcal meningitis in Africa also contribute significantly to this burden both in children and adults [2, 9,10,11].
While the diagnostic work-up typically starts with identification of classic symptoms, such as fever, headache, neck stiffness, these are largely unreliable in young children and therefore fraught with significant risk of missed diagnosis [12]. Even when ABM is associated with sepsis, appropriate treatment for sepsis may be inadequate for the accompanying ABM especially as the agents are generally different [13], hence the need for confirmatory diagnosis by the examination of cerebrospinal fluid (CSF) and identification of etiologic agent is essential to guide optimal management. Conventional methods such as Gram stain and culture have very limited value even when standard microbiology laboratories are readily available [14]. In most developing countries, microbiology diagnostic laboratories are poorly equipped, trained personnel are scarce and the turnaround time for these procedures are often suboptimal for optimizing patient care [14]. Thus, the use of non-culture based, rapid tests are needed in these settings to improve diagnostics and optimize patient management. Multiplex polymerase chain reaction (PCR) is one such approach that can improve the detection of bacterial, viral, and fungal agents in the CSF through the detection of microbial nucleic acids, including from non-viable organisms [15, 16]. Although in recent years, PCR based testing has become more readily available at national reference laboratories in several countries within the meningitis belt of Africa, these focus primarily on the detection of bacterial pathogens and are often restricted to the meningitis season [11, 16]. The aim of this study was to compare the yield from standard bacterial culture with the multiplex nested PCR platform, the BioFire® FilmArray® Meningitis/Encephalitis Panel (BioFire ME Panel). We will also aim to determine the clinical usefulness of multiplex PCR for the simultaneous detection of common pathogens associated with meningitis in archived CSF samples of suspected cases of meningitis in young children who were evaluated for community-acquired bacteremia syndromes in Nigeria in comparison with standard diagnostics. We utilized the US Food and Drug Administration (FDA) cleared multiplex PCR (mPCR) platform, the BioFire® FilmArray® Meningitis/Encephalitis Panel (BioFire ME Panel) (BioFire Diagnostics, LLC, Salt Lake City, UT) which tests for six bacteria (Escherichia coli K1, H. influenzae, Listeria monocytogenes, Neisseria meningitidis, Streptococcus agalactiae, and Streptococcus pneumoniae) seven viruses (cytomegalovirus [CMV), enterovirus [EV), herpes simplex virus type 1 [HSV-1), herpes simplex virus type 2 [HSV-2), human herpes virus type 6[(HHV-6), human parechovirus [HPeV), and varicella zoster virus [VZ))) and one yeast (Cryptococcus neoformans/gattii). Using this platform, results are generated in about an hour for each sample.
Materials and methods
Enrollment sites
The enrollment sites at Federal Capital Territory (FCT) are as previously described [17, 18]. In Kano we enrolled children from Aminu Kano Teaching Hospital, Hasiya Bayero Paediatric Hospital, and Murtala Mohammed Specialist Hospital. While Aminu Kano Teaching Hospital serves as a tertiary referral center, the other 2 facilities provide primary and secondary care services. The combined outpatient attendance for children at these 3 facilities is about 1000 daily. Kano is the capital of Kano state in northwest Nigeria and one of the oldest cities in the country. Although largely urban, Kano is not as cosmopolitan as Abuja but is more densely populated. The entire state is within the meningococcal disease belt. Malaria transmission is seasonal [19], and HIV prevalence among women attending antenatal clinic is 1.3% [20, 21].
Participant enrollment
Trained study physicians and research assistants were stationed in the emergency units, clinics, and wards of the participating hospitals from where they enrolled participants. Eligible patients were those aged less than five years and presenting with fever of > 3 days duration in addition to prostration, convulsion, refusal to feed, diarrhea, or altered consciousness were eligible. Signed informed consent was obtained from the parent or guardian. A physician administered a detailed questionnaire to obtain information on clinical history and physical examination findings.
Clinical specimen collection and initial processing
In children with a clinical suspicion of meningitis, CSF was collected through lumbar puncture in a labelled sterile vial by a medical officer under aseptic conditions and transported to the microbiology laboratory, no longer than 2 h after collection. From specimens with total volume over 2 mL, aliquots of the specimens were aseptically transferred and stored in cryovials at minus 80 °C for future diagnostic testing.
A few drops of a centrifuged portion of the sample were placed on a clean glass slide, air dried, heat fixed, Gram stained and examined for blood cells and bacteria. A loopful of the sample was inoculated onto 5% sheep blood agar, chocolate agar, and MacConkey agar. The 5% sheep blood agar, and chocolate agar plates were incubated in 5% CO2 while the MacConkey agar was incubated in ambient air. Incubation for all culture media was at 37˚C for 24 to 48 h. After incubation, bacteria were identified by their colony morphology, phenotypic tests, and limited biochemical testing such as oxidase test, indole test, citrate test and TSI tests were performed. Rapid antigen detection kits, viral or fungal culture facilities were not available and hence, these tests are not performed.
Cerebrospinal fluid specimens (n = 400) originally collected from the Community-acquired bacteremic syndromes in Young Nigerian Children (CABSYNC) and Community-acquired Pneumonia and Invasive Bacteria Disease (CAPIBD) projects were submitted for this study. In addition to these we also processed residual samples collected from patients in the northwest zone of the country during meningitis outbreaks.
Samples met inclusion criteria if they had not been centrifuged, had a residual volume of ≥ 200 μl and are not visibly blood stained. All specimens were collected between January 2014—January 2018, stored at minus 80˚C before use and did not undergo any freeze–thaw cycles prior to testing by BioFire® ME Panel.
Ethical approval
This study was reviewed and approved by the Federal Capital Territory (FCT) Ethics Committee (approval number FCTA/HHSS/NH/GEN/54/II/128) and the Ethics Review Committee of the Kano State Health Services Management Board (Approval numbers AKTH/MAC/SUB/12A/P-3/VI/972 and NHREC/21/08/2008/AKTH/EC/872).
BioFire® ME Panel testing
Approximately 200 µl of CSF was subjected to testing with the BioFire® ME Panel according to the manufacturer’s instructions. The closed FilmArray system performs nucleic acid extraction, reverse transcription, and multiplex nested PCR, automatically interprets endpoint melting curve data and provides the result in approximately 1 h. The BioFire® ME Panel tests for genetic targets from E. coli K1, H. influenzae, L. monocytogenes, N. meningitidis, S. agalactiae, S. pneumoniae, CMV, EV, HSV-1, HSV-2, HHV-6, HPeV, VZV and C. neoformans/gattii.
Each BioFire® ME panel pouch contains two internal controls; an RNA process control that targets RNA transcript from the yeast Schizosaccharomyces pombe (freeze-dried into the pouch and rehydrated upon addition of sample, which was carried through all stages of the test process) and a PCR2 control that detects a DNA target dried into the wells of the array (which ensures that the second stage PCR is successful). A run is considered successful only if the two internal controls are valid.
Upon installation of the equipment at the International Foundation Against Infectious Diseases in Nigeria (IFAIN) Central Laboratory in Abuja, five frozen external controls mixes, covering all 14 targets, were provided for daily testing. FilmArray operators completed a valid external control run on each day of specimen run.
BioFire® ME Panel performance: The procedure for testing the specimens proceeded without any hitches and was uneventful.
Verification of results from specimens with mixed bacterial agents
The AriaMx Real-Time PCR System is an integrated quantitative PCR system which performs amplification, detection, and analysis within the same equipment. It consists of inbuilt thermal cycler, an optical system with an LED excitation source and comprehensive data analysis program. The instrument holds up to six modules. The scanning optics design is able to deliver optimal separation between the dyes and the samples. The PCR system has a closed-tube detection format that can be used with a range of fluorescence detection chemistries including SYBR Green and EvaGreen dyes as well as flurogenic probe systems including Taqman probes [22, 23].
The 12 samples with mixed bacterial agents were tested using the Direct Real-Time protocol on the AriaMx [24]. This involves making a reaction mixture with 2 μl of neat CSF sample with 12 μl of Quanta Perfecta qPCR ToughMix Low ROX MasterMix, 7.5 μl of PCR grade water, and 1.0 μl each of forward primer, backward primer, and probe, giving a total reaction volume of 25 μl per well. For each of the target bacterial agents, Species-specific primers, and probes (SodC for N. meningitidis, hpd H. influenzae and LytA for S. pneumoniae) were used. Positive DNA controls for each of the target bacteria were included to validate each run, as well as No Template Controls (NTC) as negative controls to check for possible contamination.
At the end of the run, the PCR machine calculates the cycle threshold (Ct) for each gene target by determining the signal strength at which the fluorescence exceeded the threshold limit. The presence or absence of the target gene in each isolate is determined by the Ct value. Specimens with Ct value ≤ 35 were considered positive. Where the specimen yielded a valid Ct value for more than one target gene, the specimen was retested for the target gene involved and was only reported as positive if the repeat test gave the same result as the initial test.
Results
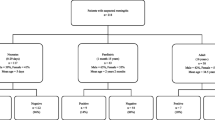
Four hundred CSF specimens consisting of 372 (93.0%) from children aged less than 5 years and 27 (6.8%) from an incidental meningococcal disease outbreak in a neighboring state within the northwest zone of Nigeria were included in the study. Over 90% of the samples were collected from Kano, where most children were enrolled from Murtala Mohammed Specialist Hospital (Table 1). Young infants aged less than 2 months constituted 55/400 (13.6%) of the participants (Table 2). Bacterial culture was positive in 32/400 (8%) of the specimens, while 368 specimens were culture negative. The distribution of agents detected from CSF bacterial culture is summarized in Fig. 1.
All 400 CSF specimens were tested with the BioFire ME Panel and positive results were obtained from 107/400 (26.75%) specimens. The BioFire ME Panel did not detect any pathogen in 293 (73.25%) samples. The BioFire ME panel detected all bacterial culture positive specimens, except Acinetobacter, Staphylococcus, and Salmonella which are not on the panel. Single viral agents were detected in 11 (2.7%) samples and a single bacterial agent was detected in 76 (19%) samples. In 10 (2.5%) samples there were mixed bacterial and viral agents detected and 12 (3.0%) samples had mixed bacterial agents detected. Overall, the frequency of pathogens detected was as follows; S. pneumoniae 42 (10.5%), N. meningitidis 32 (8%), H. influenzae 29 (7.3%), E. coli 5 (1.3%), Streptococcus agalactiae 1 (0.3%). The frequency of viral agents detected was as follows; CMV 10 (2.5%), HHV-6 6 (1.5%), VZV 3 (0.8%), HSV-1 2 (0.5%), and EV 2 (0.5%) (Fig. 1 and Table 3).
Of the 368 bacterial culture negative CSF specimens, 77 (20.9%) were positive with the BioFire ME Panel for at least one bacterial agent. S. pneumoniae was the most commonly detected bacterial pathogen by culture (n = 11) and by the BioFire ME Panel (n = 42). The age distribution of cases identified by BioFire ME Panel reveals a predominant proportion of H. influenzae in infants while N. meningitidis was more prevalent in older children (Fig. 2). BioFire ME also offers, for the first time from this setting, data on the viral agents associated with meningitis. Single viral agents were detected in 11 (2.8%) samples while co-detections with bacterial agents or other viruses occurred in 23 (5.8%) samples (Table 3).
Verification of results from specimens with mixed bacterial agents
Those CSF samples which yielded mixed bacterial agents with the BioFire ME Panel were retested using the AriaMx PCR platform as described in materials and methods. There were two samples which did not provide 100% concordance for multiple pathogens detected by BioFire ME Panel when compared to AriaMx PCR. In one, N. meningitidis and S. pneumoniae were yielded on the BioFire ME Panel but yielded only N. meningitidis on the AriaMx PCR platform while in the other, H. influenzae and N. meningitidis was detected in BioFire ME Panel and H. influenzae and S. pneumoniae detected in AriaMx PCR. All other multiple pathogens detected by BioFire ME Panel were also detected by AriaMx PCR platform provided the target primers were present in the platform. Hence, they are not analytical false positives as they were detected across multiple platforms. In some cases, the AriaMx PCR platform detected a different pathogen than the culture. This included one H. influenza that was identified as alpha-hemolytic Streptococcus, one N. meningitidis that was also identified as an alpha-hemolytic Streptococcus by culture, and another N. meningitidis that was identified as Acinetobacter species. There were no PCR targets for bacteria isolated in eight of the 32 culture positive specimen (Fig. 1).
We had primarily CSF bacterial culture results on these patients as the source facilities were not resourced to perform CSF chemistry or cell count. The acquisition of clinical outcome data was inconsistent, making it difficult to establish a comprehensive correlation between all laboratory results and patient outcomes. The documentation of clinical outcome data was limited, with most records available only during the initial days of hospitalization. Overall, there were 24 observations which included 7 fatalities (Table 4). These fatalities included two patients with dual infections with S. pneumoniae and N. meningitidis detected on both PCR panels and in one case detected by the BioFire ME Panel alone without confirmation on the AriaMx.6
Discussion
The typical clinical approach for the management of ABM relies on bacterial culture of CSF, which is widely recognized as the gold standard, even though results are often not available until 48 h or more after specimen collection. In practice, most patients are started on empiric, broad spectrum antibiotics, such as a third-generation cephalosporin, but this may not be indicated in all forms of ABM and additional adjunct therapy may be required in specific forms of meningitis, such as steroids in H. influenzae type b or S. pneumoniae. Thus, delay in definitive etiologic diagnosis of ABM compromises optimal care. Other rapid methods for pathogen detection such as Gram stain have a broad range of sensitivity and are pathogen and population dependent [14]. CSF pleocytosis has very poor specificity in guiding diagnosis of ABM in general, but particularly in young children as counts may be normal despite a diagnosis of ABM [25, 26]. For the detection of viruses, in patients who present with “aseptic meningitis”, molecular detection offers the only accurate diagnostic approach. For detection of cryptococcal meningoencephalitis, lateral flow cryptococcal antigen tests should be the first line test as numerous studies have demonstrated they are the most sensitive, particularly in the early stages of disease and when fungal burden is low. During the past several years the BioFire ME Panel has been extensively evaluated across several developed country settings and has been widely adopted for routine diagnostics for suspected cases of ME [27,28,29,30]. The minimal hands-on time for specimen processing, rapid turnaround time and the small CSF volume required for testing multiple pathogens simultaneously makes it particularly attractive for pediatric use.
Our preliminary data from central and northwest Nigeria were generated from archived CSF specimens that were collected from children who presented with severe clinical symptoms such as high fever, irritability, bulgy fontanel, or seizures. In this analysis of archived collection of CSF samples, we utilized, for the first time in this setting a multiplex PCR platform for the simultaneous detection of both viral and bacterial agents. In contrast to reports from most developed country settings, viral agents such as enterovirus was detected infrequently in our patients. Our results also contrast with observations from Ethiopia, which is also within the meningitis belt of Africa and may have had the pioneer experience of using the BioFire ME Panel. Of 218 CSF samples, 21 (10%) were PCR positive; 4% in neonates,14% in children and 18% in adults. Virus was detected in 57% of the PCR positive samples, bacteria in 33% and fungi in 10% [31]. This contrast may in part be due to the fact that our enrollment was at secondary and tertiary facilities where severely ill children are more likely to present and our criteria included children with overt clinical signs of meningitis.
While most countries have now introduced conjugate vaccines against the leading bacterial agents of ABM and have observed significant decline in the incidence of ABM caused by these agents, pneumococcal conjugate vaccine has only recently been introduced to the National Immunization Program in Nigeria and routine immunization coverage in general, remains very low in most parts of northern Nigeria, including Kano [32], the location that contributed the largest number of specimens for this study. In this report, we reveal for the first time the comparative yield from CSF of bacterial culture and PCR from children with suspected ABM in Nigeria. In addition, this also provides the first report of multiplex PCR testing of CSF obtained from Nigerian children for several viruses. The poor performance of bacterial culture, which is the current routine approach to diagnosis of ABM raises significant concerns about the large number of cases of ABM that may be receiving suboptimal care due to missed diagnosis.
The practical interpretation of a culture-negative, PCR positive CSF specimen is likely due to the high prevalence of non-prescription antibiotic use [17]. This is because while the bacterial pathogens are not viable, the genetic material which is the target of PCR can still be detected. This is particularly likely the case in our study because the children had fever for at least three days prior to enrollment and could thus were likely took antibiotics during that time. This thus may have contributed significantly to the culture-negative but PCR-positive results discovered in this study. Other practical possibilities may include patients who present very early or have lower bacterial inoculum in CSF and thus more likely to have full recovery and less complications or are these false positive tests attributable to transient bacteria DNA in the blood stream following translocation from the sinuses or nasopharynx or contamination during specimen sampling [33]. While these remain possibilities, the context in which clinical specimens were obtained from these children with an acute febrile illness and clinical suspicion for meningitis, a diagnosis of meningitis remains the most likely possibility. The retrospective nature of the current study did not permit for the formal exploration of these possibilities.
Molecular techniques are significantly more effective for establishing the etiologic diagnosis of pyogenic meningitis, although they cannot completely replace conventional tests given the need for bacteria isolation and susceptibility testing for the determination of optimal antibiotic choice for clinical management [34]. All the patients in our series were treated empirically with ceftriaxone once a CSF sample was obtained. While this agent is a good choice for most bacterial agents that are associated with meningitis, such as S. pneumoniae, Salmonella spp. and N. meningitidis, obtaining prompt information on the etiologic agent would have modified duration of treatment for cases of N. meningitidis, which can be treated optimally with a shorter course than S. pneumoniae or H. influenzae. In addition, such information would have prompted the initiation of prophylaxis for household contacts for those children with N. meningitidis or prompted evaluation for alternative cause of illness or modification of empiric treatment.
The detection of viral agents by PCR clearly has a strong indication for initiating specific treatment for HSV and supportive care for enterovirus. However, detection of CMV and VZV or HHV6 warrant careful clinical correlation, since these are herpes viruses and detection in CSF may be secondary to reactivation rather than primary infection. The clinical relevance of these agents in this setting where co-morbid conditions such as severe malnutrition and HIV are prevalent warrants further investigation.
There are several limitations for this study. We determined the performance of the BioFire ME Panel on archived CSF specimens, relative to bacterial culture alone. We did not have a local diagnostic platform for the detection of viral and fungal agents that are included in the BioFire ME Panel. While our storage conditions for the CSF are deemed optimal, we are unable to guarantee absence of DNA degradation following prolonged specimen storage; although there are reports in the literature of specimens in storage for over three decades been screened for bacterial DNA yielding meaningful results [35, 36].
We detected a high prevalence of mixed bacterial meningitis in our population. The clinical setting where the patients are triaged and specimens collected are often congested and facemasks are not typically worn during these procedures, thus raising the likelihood of contamination at the point of collection. Mixed bacterial infections are frequently reported following cranial trauma, surgical intervention or in nosocomial infections. Nevertheless, there have been several reports of community acquired ABM with mixed bacterial agents in immunocompetent patients [37, 38]. Most frequently reported are infections caused by a combination of common meningeal pathogens such as N. meningitidis and S. pneumoniae or N. meningitidis and H. influenzae. Most of the reports in the literature of polymicrobial ABM predates the widespread use of antibiotics, with prevalence ranging from 1–4%, predominantly affecting young children and the most frequent bacteria being H. influenzae [39, 40].
In a recent report of the evaluation of CSF samples from patients with pyogenic meningitis by high-throughput sequencing of 16S rDNA, 61.5% of specimens had mixed pathogens [33]. While this report represents one of the earliest uses of multiplex sequencing in evaluating pyogenic meningitis, the results challenge the current norm of associating pyogenic meningitis with single agents that are identifiable only by culture. Where clinical outcome data is available, these polymicrobial ABM have been associated with high case fatality and neurological sequelae [40]. We were unable to completely exclude the possibility of specimen contamination at the point of collection or specimen aliquoting in the laboratory. It is less likely however, that the specimens were contaminated during BioFire ME Panel processing, since the system is self-contained, and the testing pouch is not open at any time after sample/buffer addition. We further minimized the risk of amplicon contamination by ensuring that operators wore a facemask and preparations were performed in a safety hood. Because the specimens mainly constituted a convenience sample that wasn't drawn from a representative population, drawing conclusions about the varying frequencies of infections caused by different pathogens or the notable proportion of bacterial coinfections identified becomes challenging and will not be accurately deduced from this study.
The BioFire ME panel offers a user-friendly and rapid approach for diagnosis but the cost per test will be unaffordable in low- and middle-income country settings. In the meningitis belt of Africa where outbreaks of meningococcal disease occur, these have been frequently associated with concurrent mini outbreaks of S. pneumoniae meningitis and in 2016 there was a predominant outbreak caused by S. pneumoniae serotype 1 [41]. The scarcity of diagnostic laboratories in this region makes accurate diagnosis a challenge [12, 41] While we have limited outcome data, it is plausible that in this setting, mixed bacterial meningitis may be contributing significantly to adverse outcomes, and these are underestimated due to poor diagnostics [42]. Interpretation of these results would have been significantly enhanced by a full complement of CSF analytics that include cytology and chemistry. These were not consistently available at our clinical sites. We are now planning a prospective study in this setting to evaluate the short- and medium-term impact of polymicrobial meningitis.
In conclusion, we have provided novel data on the etiologic yield for multiple pathogens detected simultaneously from children with clinical suspicion of meningitis from Nigeria, the most populous country within the meningococcal disease belt and in sub-Saharan Africa. The BioFire ME Panel detected several pathogens that were missed by culture. While the BioFire ME Panel cannot replace conventional laboratory studies and the targets can in fact be further customized for agents commonly associated with ABM in this region, the ease and rapidity of performing this test, offers a unique opportunity to improve our understanding of the epidemiology of meningitis and meningoencephalitis in this setting. Future studies integrating unbiased metagenomics pathogen sequencing CSF chemistry and cytology are planned. These studies will provide novel data that will inform the design of customized ME panels for this region. However, to maximize the deployment of such panels, they would need to be affordable with a positive cost–benefit margin in these settings where meningitis is still highly prevalent and such diagnostic devices are most needed.
Availability of data and materials
All data is available within this manuscript.
References
Kim KS. Acute bacterial meningitis in infants and children. Lancet Infect Dis. 2010;10(1):32–42. Available from: https://pubmed.ncbi.nlm.nih.gov/20129147/. [cited 2023 Apr 15].
Peltola H. Burden of meningitis and other severe bacterial infections of children in Africa: implications for prevention. clinical infectious diseases. 2001;32(1):64–75. Available from: https://academic.oup.com/cid/article/32/1/64/310637. [cited 2023 Apr 15].
Agrawal S, Nadel S. Acute bacterial meningitis in infants and children: epidemiology and management. Paediatr Drugs. 2011;13(6):385–400 Available from: https://pubmed.ncbi.nlm.nih.gov/21999651/. [cited 2023 Apr 15].
Chang CJ, Chang WN, Huang LT, Huang SC, Chang YC, Hung PL, et al. Bacterial meningitis in infants: the epidemiology, clinical features, and prognostic factors. Brain Dev. 2004;26(3):168–75. Available from: https://pubmed.ncbi.nlm.nih.gov/15030905/. [cited 2023 Apr 15].
De Louvois J, Halket S, Harvey D. Neonatal meningitis in England and Wales: sequelae at 5 years of age. Eur J Pediatr. 2005;164(12):730–4. Available from: https://pubmed.ncbi.nlm.nih.gov/16136294/. [cited 2023 Apr 15].
John CC, Carabin H, Montano SM, Bangirana P, Zunt JR, Peterson PK. Global research priorities for infections that affect the nervous system. Nature. 2015;527(7578):S178. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4697933/. [cited 2023 Apr 15].
Van De Beek D, Brouwer MC, Thwaites GE, Tunkel AR. Advances in treatment of bacterial meningitis. Lancet. 2012;380(9854):1693–702. Available from: https://pubmed.ncbi.nlm.nih.gov/23141618/. [cited 2023 Apr 15].
Harrison LH, Trotter CL, Ramsay ME. Global epidemiology of meningococcal disease. Vaccine. 2009;27(SUPPL 2):B51.
Delrieu I, Yaro S, Tamekloé TAS, Njanpop-Lafourcade BM, Tall H, Jaillard P, et al. Emergence of epidemic neisseria meningitidis serogroup X meningitis in Togo and Burkina Faso. PLoS One. 2011;6(5):e19513. Available from: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0019513. [cited 2023 Apr 15].
Traoré Y, Njanpop-Lafourcade B, Adjogble K, Lourd M, Yaro S. The rise and fall of epidemic neisseria meningitidis serogroup W135 meningitis in Burkina Faso, 2002–2005. Clin Infect Dis. 2006;43(7):817–22. Epub 2006 Aug. 22.
Chow J, Uadiale K, Bestman A, Kamau C, Caugant DA, Shehu A, et al. Invasive meningococcal meningitis serogroup c outbreak in Northwest Nigeria, 2015 - Third Consecutive Outbreak of a New Strain. PLoS Curr. 2016;8. Available from: https://pubmed.ncbi.nlm.nih.gov/27508101/. [cited 2023 Apr 15].
Weber MW, Herman J, Jaffar S, Usen S, Oparaugo A, Omosigho C, et al. Clinical predictors of bacterial meningitis in infants and young children in The Gambia. Trop Med Int Health. 2002;7(9):722–31. Available from: https://pubmed.ncbi.nlm.nih.gov/12225501/. [cited 2023 Apr 15].
Medugu N, Iregbu K, Tam PYI, Obaro S. Aetiology of neonatal sepsis in Nigeria, and relevance of group b streptococcus: a systematic review. PLoS One. 2018;13(7):e0200350.
Berkley JA, Mwangi I, Ngetsa CJ, Mwarumba S, Lowe BS, Marsh K, et al. Diagnosis of acute bacterial meningitis in children at a district hospital in sub-Saharan Africa. Lancet. 2001;357(9270):1753–7. Available from: https://pubmed.ncbi.nlm.nih.gov/11403812/. [cited 2023 Apr 15].
Başpınar EÖ, Dayan S, Bekçibaşı M, Tekin R, Ayaz C, Deveci Ö, et al. Comparison of culture and PCR methods in the diagnosis of bacterial meningitis. Braz J Microbiol. 2017;48(2):232–6. Available from: https://pubmed.ncbi.nlm.nih.gov/27793541/. [cited 2023 Apr 15].
Meyer T, Franke G, Polywka SKA, Lütgehetmann M, Gbadamosi J, Magnus T, et al. Improved detection of bacterial central nervous system infections by use of a broad-range PCR assay. J Clin Microbiol. 2014;52(5):1751. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3993695/. [cited 2023 Apr 15].
Obaro S, Lawson L, Essen U, Ibrahim K, Brooks K, Otuneye A, et al. Community acquired bacteremia in young children from central Nigeria—a pilot study. BMC Infect Dis. 2011;11:137.
Medugu N, Tickler IA, Duru C, Egah R, James AO, Odili V, et al. Phenotypic and molecular characterization of beta-lactam resistant Multidrug-resistant Enterobacterales isolated from patients attending six hospitals in Northern Nigeria. Sci Rep. 2023;13(1). Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10293160/. [cited 2023 Aug 6].
Olayemi K, Ande AT, Ayanwale AV, Mohammed AZ, Bello TM, Idris B, et al. Seasonal trends in epidemiological and entomological profiles of malaria transmission in North Central Nigeria. Pak J Biol Sci. 2011;14(4):293–9. Available from: https://pubmed.ncbi.nlm.nih.gov/21870632/. [cited 2023 Apr 15].
Awofala AA, Ogundele OE. HIV epidemiology in Nigeria. Saudi J Biol Sci. 2018;25(4):697. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5937013/. [cited 2023 Apr 15].
Odimegwu CO, Akinyemi JO, Alabi OO. HIV-Stigma in Nigeria: review of research studies, policies, and programmes. AIDS Res Treat. 2017;2017. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5763061/. [cited 2023 Apr 15].
Papavasileiou K, Papavasileiou E, Tzanakaki G, Voyatzi A, Kremastinou J, Chatzipanagiotou S. Acute bacterial meningitis cases diagnosed by culture and PCR in a children’s hospital throughout a 9-year period (2000–2008) in Athens, Greece. Mol Diagn Ther. 2011;15(2):109–13. Available from: https://link.springer.com/article/10.1007/BF03256400. [cited 2023 May 2].
Leber AL, Everhart K, Balada-Llasat JM, Cullison J, Daly J, Holt S, et al. Multicenter evaluation of biofire filmarray meningitis/encephalitis panel for detection of bacteria, viruses, and yeast in cerebrospinal fluid specimens. J Clin Microbiol. 2016;54(9):2251–61. Available from: https://journals.asm.org/doi/10.1128/JCM.00730-16. [cited 2023 May 2].
van de Beek D, de Gans J, Spanjaard L, Weisfelt M, Reitsma JB, Vermeulen M. Clinical features and prognostic factors in adults with bacterial meningitis. 2004;351(18):1849–59. Available from: https://www.nejm.org/doi/full/10.1056/nejmoa040845. [cited 2023 Apr 15].
Domingues RB, Santos MV dos, Leite FBV de M, Senne C. FilmArray Meningitis/Encephalitis (ME) panel in the diagnosis of bacterial meningitis. Braz J Infect Dis. 2019;23(6):468–70.
Tansarli GS, Chapin KC. Diagnostic test accuracy of the BioFire® FilmArray® meningitis/encephalitis panel: a systematic review and meta-analysis. Clin Microbiol Infect. 2020;26(3):281–90. Google Search. Available from: https://www.google.com/search?q=Tansarli+GS%2C+Chapin+KC%2C+Diagnostic+test+accuracy+of+the+BioFire%C2%AE+FilmArray%C2%AE+meningitis%2Fencephalitis+panel%3A+a+systematic+review+and+meta-analysis.+Clin+Microbiol+Infect+2020+Mar%3B26(3)%3A281-290.&oq=Tansarli+GS%2C+Chapin+KC%2C+Diagnostic+test+accuracy+of+the+BioFire%C2%AE+FilmArray%C2%AE+meningitis%2Fencephalitis+panel%3A+a+systematic+review+and+meta-analysis.+Clin+Microbiol+Infect++2020+Mar%3B26(3)%3A281-290.&aqs=chrome..69i57.326j0j7&sourceid=chrome&ie=UTF-8&google_abuse=GOOGLE_ABUSE_EXEMPTION%3DID%3Df04d1f531356edac:TM%3D1681564437:C%3Dr:IP%3D102.88.34.118-:S%3D-aNrp2xpufxVMPkOnEIG9m4%3B+path%3D/%3B+domain%3Dgoogle.com%3B+expires%3DSat,+15-Apr-2023+16:13:57+GMT. [cited 2023 Apr 15].
Boudet A, Pantel A, Carles M-J, Boclé H, Charachon S, Enault C, et al. A review of a 13-month period of FilmArray Meningitis/Encephalitis panel implementation as a first-line diagnosis tool at a university hospital. PLoS One. 2019;14(10):e0223887. Google Search . Available from: https://www.google.com/search?q=Boudet+A%2C+Pantel+A%2C+Carles+M-J%2C+Bocl%C3%A9+H%2C+Charachon+S%2C+Enault+C%2C+et+al.+(2019)+A+review+of+a+13-month+period+of+FilmArray+Meningitis%2FEncephalitis+panel+implementation+as+a+first-line+diagnosis+tool+at+a+university+hospital.+PLoS+ONE+14(10)%3A+e0223887.&oq=Boudet+A%2C+Pantel+A%2C+Carles+M-J%2C+Bocl%C3%A9+H%2C+Charachon+S%2C+Enault+C%2C+et+al.+(2019)+A+review+of+a+13-month+period+of+FilmArray+Meningitis%2FEncephalitis+panel+implementation+as+a+first-line+diagnosis+tool+at+a+university+hospital.+PLoS+ONE+14(10)%3A+e0223887.&aqs=chrome..69i57.440j0j7&sourceid=chrome&ie=UTF-8&google_abuse=GOOGLE_ABUSE_EXEMPTION%3DID%3Df6aca2a9b2d6da26:TM%3D1681564440:C%3Dr:IP%3D102.88.34.118-:S%3DM1r4dSFkZSreEBUbzEFj3xk%3B+path%3D/%3B+domain%3Dgoogle.com%3B+expires%3DSat,+15-Apr-2023+16:14:00+GMT. [cited 2023 Apr 15].
Nabower AM, Miller S, Biewen B, Lyden E, Goodrich N, Miller A, et al. Association of the filmarray meningitis/encephalitis panel with clinical management. Hosp Pediatr. 2019;9(10):763–9. Available from: https://pubmed.ncbi.nlm.nih.gov/31511395/. [cited 2023 Apr 15].
O’Brien MP, Francis JR, Marr IM, Baird RW. Impact of cerebrospinal fluid multiplex assay on diagnosis and outcomes of central nervous system infections in children: a before and after cohort study. Pediatr Infect Dis J. 2018;37:868–71. Google Search. Available from: https://www.google.com/search?q=O%27Brien+MP%2C+Francis+JR%2C+Marr+IM%2C+Baird+RW+(2018)+Impact+of+Cerebrospinal+Fluid+Multiplex+Assay+on+Diagnosis+and+Outcomes+of+Central+Nervous+System+Infections+in+Children%3A+A+Before+and+After+Cohort+Study.+The+Pediatric+Infectious+Disease+Journal+37%3A868%E2%80%93871.&oq=O%27Brien+MP%2C+Francis+JR%2C+Marr+IM%2C+Baird+RW+(2018)+Impact+of+Cerebrospinal+Fluid+Multiplex+Assay+on+Diagnosis+and+Outcomes+of+Central+Nervous+System+Infections+in+Children%3A+A+Before+and+After+Cohort+Study.+The+Pediatric+Infectious+Disease+Journal+37%3A868%E2%80%93871.&aqs=chrome..69i57.315j0j7&sourceid=chrome&ie=UTF-8&google_abuse=GOOGLE_ABUSE_EXEMPTION%3DID%3Dd29faefb64e3767a:TM%3D1681564475:C%3Dr:IP%3D102.88.34.118-:S%3D01ykEnLpHfOAtj89CccyBL0%3B+path%3D/%3B+domain%3Dgoogle.com%3B+expires%3DSat,+15-Apr-2023+16:14:35+GMT. [cited 2023 Apr 15].
Cailleaux M, Pilmis B, Mizrahi A, Lourtet-Hascoet J, Van JCN, Alix L, et al. Impact of a multiplex PCR assay (FilmArray®) on the management of patients with suspected central nervous system infections. Eur J Clin Microbiol Infect Dis. 2019;39(2):293–7. Available from: https://europepmc.org/article/med/31720944. [cited 2023 Apr 15].
Bårnes GK, Gudina EK, Berhane M, Abdissa A, Tesfaw G, Abebe G, et al. New molecular tools for meningitis diagnostics in Ethiopia - a necessary step towards improving antimicrobial prescription. BMC Infect Dis. 2018;18(1). Available from: https://pubmed.ncbi.nlm.nih.gov/30572843/. [cited 2023 Apr 15].
Olorunsaiye CZ, Degge H. Variations in the uptake of routine immunization in nigeria: examining determinants of inequitable access. 101080/2376200420161206780 [Internet]. 2016;2(1):19–29. Available from: https://www.tandfonline.com/doi/abs/10.1080/23762004.2016.1206780. [cited 2023 Apr 15].
Liu A, Wang C, Liang Z, Zhou ZW, Wang L, Ma Q, et al. High-throughput sequencing of 16S rDNA amplicons characterizes bacterial composition in cerebrospinal fluid samples from patients with purulent meningitis. Drug Des Devel Ther. 2015;9:4417. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4535540/. [cited 2023 Apr 15].
Hanson KE. The first fully automated molecular diagnostic panel for meningitis and encephalitis: how well does it perform, and when should it be used? J Clin Microbiol. 2016;54(9):2222. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5005496/. [cited 2023 Apr 15].
Jerome KR, Huang ML, Wald A, Selke S, Corey L. Quantitative stability of DNA after extended storage of clinical specimens as determined by real-time PCR. J Clin Microbiol. 2002;40(7):2609–11. Available from: https://pubmed.ncbi.nlm.nih.gov/12089286/. [cited 2023 Apr 15].
Chang WN, Lu CH, Huang CR, Chuang YC. Mixed infection in adult bacterial meningitis. Infection. 2000;28(1):8–12. Available from: https://pubmed.ncbi.nlm.nih.gov/10697784/. [cited 2023 Apr 15].
Corless CE, Guiver M, Borrow R, Edwards-Jones V, Fox AJ, Kaczmarski EB. Simultaneous detection of neisseria meningitidis, haemophilus influenzae, and streptococcus pneumoniae in suspected cases of meningitis and septicemia using real-time PCR. J Clin Microbiol. 2001;39(4):1553. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC87969/. [cited 2023 Apr 15].
Downs NJ, Hodges GR, Taylor SA. Mixed bacterial meningitis. Rev Infect Dis. 1987;9(4):693–703. Available from: https://academic.oup.com/cid/article/9/4/693/279408. [cited 2023 Apr 15].
Herweg JC, Middelkamp JN, Hartmann AF. Simultaneous mixed bacterial meningitis in children. J Pediatr. 1963;63(1):76–83. Available from: https://pubmed.ncbi.nlm.nih.gov/13954177/. [cited 2023 Apr 15].
Gessner BD, Mueller JE, Yaro S. African meningitis belt pneumococcal disease epidemiology indicates a need for an effective serotype 1 containing vaccine, including for older children and adults. BMC Infect Dis. 2010;10(1):1–10 Available from: https://bmcinfectdis.biomedcentral.com/articles/10.1186/1471-2334-10-22. [cited 2023 Apr 15] .
Lehmann D, Yeka W, Rongap T, Javati A, Saleu G, Clegg A, et al. Aetiology and clinical signs of bacterial meningitis in children admitted to Goroka Base Hospital, Papua New Guinea, 1989–1992. Ann Trop Paediatr. 1999;19(1):21–32. Available from: https://pubmed.ncbi.nlm.nih.gov/10605517/. [cited 2023 Apr 15].
Precit MR, Yee R, Pandey U, Fahit M, Pool C, Naccache SN, et al. Cerebrospinal fluid findings are poor predictors of appropriate filmarray meningitis/encephalitis panel utilization in pediatric patients. J Clin Microbiol. 2020;58(3). Available from: https://journals.asm.org/doi/10.1128/JCM.01592-19. [cited 2023 Apr 15].
Acknowledgements
Our gratitude goes to the entire CAPIBD and CABSYNC teams who worked tirelessly across all sites to make sure the studies succeed.
Disclaimer
The findings and conclusions contained within are those of the authors and do not necessarily reflect positions or policies of the funders. The funders had no role in study design, data collection and analysis, or preparation of the manuscript.
Funding
SO received funding from BioFire Diagnostics/bioMérieux.
Author information
Authors and Affiliations
Contributions
SO conceived the study. NM, GO, CD conducted laboratory analyses. SO, NM conducted the scientific and statistical analyses. SO, NM, GO wrote the manuscript. SO, NM, GO, FH, BJ, SG, VO, AJ, CD, BE critically reviewed the manuscript. All authors provided critical feedback and helped shape the research, analysis, and manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The ethical review boards of Aminu Kano Teaching Hospital, Hasiya Bayero Pediatric Hospital, and Murtala Mohammed Specialist Hospital, Federal Capital Territory of Nigeria Ethics Committee, Zankli Medical Center Ethics Committee, National Hospital Abuja Ethics Committee, and Keffi Medical Center Ethics Committee reviewed and approved the study. Written informed consent was obtained from the parent or guardian of all the children in the study. All study protocols were in accordance with the Declaration of Helsinki. All data/samples were fully anonymized.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Obaro, S., Hassan-Hanga, F., Medugu, N. et al. Comparison of bacterial culture with BioFire® FilmArray® multiplex PCR screening of archived cerebrospinal fluid specimens from children with suspected bacterial meningitis in Nigeria. BMC Infect Dis 23, 641 (2023). https://doi.org/10.1186/s12879-023-08645-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-023-08645-7