Abstract
Background
Malaria cases in non-endemic zero-indigenous case areas are most likely to have been imported whatever of the route of importation. In countries recently declared malaria-free and now without local transmission, imported cases remain a threat to re-introduction of the disease and a burden on the health system.
Case presentation
Three days after returning from a long trip to malaria- endemic countries; Abyei-Sudan, Chad and Uganda, a 41-year-old male resident from Jericho, Palestine, suffered paroxysms of fever, general fatigue, myalgia, arthralgia, headache, and a strong desire to vomit. Thin and thick Giemsa-stained blood smears were prepared and examined microscopically using oil immersion. Immature trophozoites (ring forms) were seen to parasitize approximately 10% of the erythrocytes revealing hyperparasitemia equivalent to > 100,000 parasites/ µl indicating severe malaria [1, 2]. The double chromatin configuration (headphones) and accolé (applique) position are both indicative of Plasmodium falciparum infection. The 18S rRNA- PCR targeting the rPLU6-rPLU5 region was used to confirm the diagnosis. The next-generation sequencing (NGS) method was carried out according to the manufacturer’s instructions (Illumina® DNA Prep, (M) Tagmentation kit (20060060), Illumina) to identify Plasmodium spp. Furthermore, NGS produced a whole-genome sequence of 22.8Mbp of the 14 chromosomes and 25Kbp of the apicoplast. A BLAST search of the apicoplast DNA and selected chromosomal DNA revealed that P. falciparum was the causative agent. The merozoite surface protein-1 (msp-1) was used to construct a phylogenetic tree of 26 P. falciparum, including the one isolated from the patient from Jericho, which clustered with the Sudanese isolate indicating genetic relatedness between the two.
Conclusion
The travel history together with signs and symptoms of malaria, followed by prompt diagnosis using conventional microscopic inspection of Giemsa-stained films together with molecular DNA tracking tools like msp-1 were key means in tracking the place of origin of infection in the case of travel to multiple destination.
Similar content being viewed by others
Background
Malaria is a potentially lethal infection caused by an apicomplexan parasite of the genus Plasmodium. It is a vector-born disease transmitted by female mosquitoes of the genus Anopheles. Of the four clinicallysignificant species in the genus, Plasmodium falciparum is the most widely distributed in Africa and having the most severe clinical consequences [3, 4]. The global burden of malaria, morbidity and mortality, is considered to be the highest among all the parasitic diseases with 247 million cases and 619,000 deaths per annum [4]. Sub-Saharan Africa is the geographical region affected most bearing 95% of the deaths with 50% of them occurring in the following four African countries, Nigeria, The Democratic Republic of Congo, Uganda, and Mozambique [4].With exception of Saudi Arabia and Yemen, all the Middle Eastern countries are free of endogenous cases of malaria, however, with most of these countries reporting imported cases [5]. At the turn of the 20th century, Palestine was described as one of the most highly malarious countries in the world [6]. Reports as early as 1880 confirmed that malaria caused by P. falciparum and P. vivax was a major public health problem. The malaria infection rate ranged from 50, 20, 7, and 0.7% in the years 1905, 1913, 1922, and 1944, respectively [7,8,9]. Six to eight species of anopheline mosquitoes were collected as larvae and adults from Palestinian marshes, swamps, and springs [6, 10]. Several attempts were conducted to eliminate malaria, starting during Ottoman rule, then during British and Jordanian rule that led to the eradication of the disease by end of the 1960 [6, 8, 10, 11]. According to International Classification of Diseases for Morbidity and Mortality Statistics, 11th Revision, v2022-02 (ICD11) a disease caused by P. falciparum is characterized by fever, chills, headache, myalgia, arthralgia, weakness, nausea, anemia, jaundice, vomiting, or diarrhea. Other more severe manifestations include splenomegaly, hypoglycemia, pulmonary or renal dysfunction, and neurological changes such as mental confusion and seizures followed by coma and death. Here the geographical origin of a rare case of malaria imported into Palestine was tracked using next-generation sequencing and population genetics.
Case presentation
A 41-year- old Palestinian man from Jericho worked as an engineer with an international institution in Abyei, a disputed area between Sudan and South Sudan. In 2018, he moved from Abyei to Eastern Chad and, spent a few days on an official mission. Then, he flew to Kampala, the capital of Uganda, where he spent one week at the institution headquarters and finally, flew home to Palestine through Dubai and Amman, Jordan. Three days after reaching his final destination in Jericho, Palestine, he started to suffer a high fever with chills and sweating. These paroxysms of fever, chills and rigor were accompanied by general fatigue, a headache and strong need to vomit despite not having eaten since the start of the symptoms. In addition, he suffered myalgia and arthralgia. He was admitted to the Jericho Government Hospital for a clinical follow up where his clinical history revealed incompliance with the recommended prophylactic treatment when using Malarone® (GlaksoSmithKine, GSK). Urinalysis, stool analysis, complete blood count, and examination of blood films were done, all of which were normal. Thick and thin blood films were prepared and examined in between the paroxysms. Blood films were stained for one hour with Giemsa’s stain made with 1:40 phosphate buffered water according to Garcia [12]. Blood films revealed the presence of the parasitaemia seen as immature trophozoites (ring forms) and indicating malaria (Fig. 1a). According to World Health Organization (WHO) malaria grading system, the case was considered hyperparasaetimic owing to the presence of >10 infected RBCs per 100 normal RBCs in an oil immersion field equivalent to > 100,000 parasites/µl indicating severe malaria (Fig. 1a). In some maturing trophozoites ring forms with thicker cytoplasm were seen occasionally in stained thin smears. DNA was extracted from the stained smears, using a QIAamp® DNA Mini Kit according to manufacturer’s instructions (Qiagen, GmbH, Hilden, Germany). Genomic DNA was used for PCR amplification, according to Snounnou et al., which amplified the 18S rRNA gene [13]. Briefly, in 25-µl-reaction; was conducted, 0.25µM of each of the rPLU6/rPLU5 primer pair, in a Syntezza Bioscience reaction tube (Jerusalem). A known Plasmodium falciparum DNA (Provided by Prof. Ron Dzikowski), 0.04ng/µl (1ng/25µl reaction) of DNA and PCR grade water was used as negative control. A PCR product of approximately 1200 bp was obtained from the DNA of the infecting parasites, which was equivalent to the positive control after running the samples on a 2% agarose gel and staining with ethidium bromide, using the Biorad gel doc XR documentation system (Bio-Rad Laboratories Inx, USA) (Fig. 1b). In addition to confirm the positive results of the blood films and determine the PCR was meant to assess suitability, the quantity and quality of the DNA for the WGS.
(a) A thin blood film made from blood from the case of malaria and stained with Giemsa’s stain as seen microscopically at X1000 magnification. Red numbers indicate erythrocytes infected with immature trophozoites (ring form) of Plasmodium spp. Erythrocyte 3 shows the double chromatin configuration (headphones) and erythrocyte 4 is in the accolé (applique) position, both of which are indicative of an infection of Plasmodium spp.(b) An agarose gel showing the amplification of region rPLU6-rPLU5, the region of the 18S rRNA gene, showing: 1, the 100 bp marker; 2 and 3, two samples, one after one week and the other after two weeks post-treatment; 4, pre-treatment sample during the acute phase; 5 and 6, negative and positive controls, respectively
The WGS analysis exposed 14 chromosome (22.8Mbp) and an apicoplast (25Kbp) DNA sequences [14]. The whole genome sequence (Accession numbers CP101623-36) and apicoplast sequence (Accession number OP661153) were deposited in the GenBank. Using a BLAST search, the apicoplast DNA,msp-1 (Accession number OP115966), msp-2 (Accession number OR198912), and any selected chromosomal DNA showed that P. falciparum caused the imported case of malaria. Based on the WHO recommendations and other studies, population genetics of the genus Plasmodium was based on the hypervariable region (556bp) of the merozoite surface protein I (msp-1) owing to its extensive polymorphism, stability throughout the life cycle, the availability of the widely used data and the easy distinguishable [15,16,17]. The Jericho patient msp-1 sequence was retrieved from the WGS resulting from this report. The phylogenetic tree based on the msp-1 of 26 isolates of P. falciparum from different countries was constructed. This include one from the resident of Jericho. The tree showed that this isolate clustered with the Sudanese isolate (Fig. 2). In accordance with the solid laboratory diagnosis of malaria caused by P. falciparum, the patient was treated for three days with tablets (artemether-lumefantrine) of the antimalarial drug Coartem® (Novartis) as the drug of choice for acute malaria. The patient ingested four tablets every eight hours on day one, and four tablets every 12 h on days two and three. His condition improved with immediate resolution of all the acute symptoms, which was confirmed on examining thin blood films and employing PCR testing (Fig. 1b) one week and two weeks post-treatment.
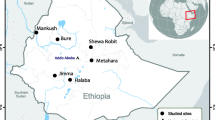
UPGMA phylogenetic tree was constructed using MEGA-X free software, and showing relationships between the sequence of the msp-1 DNA of the isolate of the P. falciparum imported into Jericho and the 25 msp-1 DNA sequences from other strains of Plasmodium retrieved from the GenBank. Plasmodium spp. msp-1 DNA sequence isolated from gorilla clade G3 (Accession number LT969558.1) was used as an out-group. A bootstrap value of 1000 replicates was used to ensure robustness of the tree and shown as percentages at tree nodes. The tree was drawn to scale with branch lengths given in the same units as those of the evolutionary distances used to infer the phylogenetic tree [18,19,20,21]. The map shows the patient’s travel through five different geographical regions, two to three weeks before the appearance of the symptoms suggestive of malaria. Map was generated with Epi Info™ CDC-free software
Discussion
This quite severe case of malaria was diagnosed in Jericho, Palestine, a zero-indigenous case area and declared by the WHO as a country free of malaria transmission [22], which made it difficult to decide the exact etiology based solely on intermittent high fever. Other diseases in the area causing fever, e. g. visceral leishmaniases, meningitis, pneumonia, malignant tumors, and viral hepatitis, were possible suspects. The patient’s travel history to and in African, malaria-endemic regions, followed by episodes of fever made malaria highly suspect and examining stained blood films followed by employing PCR and NGS methods confirmed malaria caused by P. falciparum, the most severe form of malaria. This proved the efficacy of using NGS and msp-1 DNA in tracking the geographical origin of the parasites seen in the stained smears. The main limitation was the need of a high depth of sequencing coverage (6.2x) to cover the WGS of P. falciparum (22.8Mbp), which is within a much larger amount of human host DNA (3.2Gbp). Regarding this case, local transmission was ruled out owing to the lengthy incubation period of malaria caused by P. falciparum of at least two weeks. Also, in theory, local transmission could not have occurred, owing to the absence of the main vectors of P. falciparum, i. e., Anopheles gambiae and Anopheles funestus, as evidenced by the complete absence of anopheline species during insect trapping campaigns in Palestine in the last two decades [23,24,25]. A possibility of transmission is through airport malaria, in which a traveler hopped between several airports, including ones in countries where malaria is endemic (Fig. 2). There have also been reports of ‘baggage malaria’ where anopheline mosquitoes find their way into and surviving in luggage and goods being transported from African countries where malaria is endemic to countries devoid of malaria and transmitting malaria there [26,27,28,29,30]. The other possibility, as is the situation described in this article, the patient is an extensive traveler [31]. In this case, the clinical history ruled out the further routes of transmission, e. g., such as blood transfusion, nosocomial (hospital-acquired) infection, post-transplant, parenteral such as drug abuse needle sharing, being immuno-compromised, and recrudescence (relapsing) after having had malaria [32, 33]. In theory, the patient could have contracted his infection in one of the three African countries he visited Sudan, Chad, and Uganda. Also, airports in these countries and those of Dubai and Amman may have been the venue of his infection. Population genetics was used to track the origin of his infection by correlating the DNA of P. falciparum extracted from his infected blood with DNA from reference strains from other countries around the world. The phylogenetic tree based on the msp-1 of all the samples used clustered his causative agent with a Sudanese one, indicating, most likely, that the patient contracted his infection in the Abyei region of Sudan and not from any other region he travelled to. Imported cases of malaria from regions of malaria endemicity to those of non-endemicity are common. From 1990 and early 2021, 35 cases of malaria were recorded in Palestine but without mentioning whether the diagnosis was only symptom-based or also laboratory-confirmed, or if these cases were imported or autochthonous [34]. In 2005, the Palestinian Ministry of Health declared that it has completely controlled many infections including malaria [34]. Ten years later, the WHO declared Palestine a country free of local transmission of malaria [22]. The case imported into Jericho and described here and whole-genome sequenced, permitting species identification and tracking of the cause and origin of the infection, is the only one dealt in this way in Palestine so far.
Conclusion
Cases of malaria imported into non-endemic countries, Palestine, is a challenge to their health system’s preparedness to diagnose and treat such cases. The careful taking of clinical histories, laboratory confirmation of cases including the identification of the infectious agents, using microscopy and molecular-based methods, and tracking the origin of the infection are recommended to enable timely life-saving treatment and to understand the disease epidemiology. Travelers to and from endemic countries should be dealt with according to WHO international health regulations and their amendments [35]. Monitoring for the presence of species of Anopheles in recently declared malaria free areas is crucial in preventing re-introduction of the disease.
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- PCR:
-
Polymerase chain reaction
- NGS:
-
Next-generation sequencing
- WGS:
-
Whole-genome sequence
- msp-1 :
-
Merozoite surface protein-1
- UPGMA:
-
Unweighted Pair Group Method with Arithmetic Mean
- RBC:
-
Red blood cell
References
WorldHealthOrganization. Malaria Parasite Counting. Malaria Microscopy Standard Operating Procedure- MM-SOP-09. In: WHO/HTM/GMP/MM/SOP/201609.2016.
Blood Specimens - Microscopic Examination. [https://www.cdc.gov/dpdx/diagnosticprocedures/blood/microexam.html#print].
Ashley EA, Pyae Phyo A, Woodrow CJ. Malar Lancet. 2018;391(10130):1608–21.
WorldHealthOrganization. : World malaria report 2022. In. Geneva; 2022 Licence: CC BY-NC-SA 3.0 IGO.
Al-Awadhi M, Ahmad S, Iqbal J. Current status and the epidemiology of Malaria in the Middle East Region and Beyond. Microorganisms 2021, 9(2).
Austen EE. Anti-mosquito measures in Palestine during the campaigns of 1917–1918. Trans Roy Soc Trop Med Hyg. 1919;13(4):47–60.
Cropper J. The Malarial Fevers of Palestine and their Prevention. J Royal Inst Public Health. 1907;15(2):95–9.
Kalbian VV. Jerusalem in Malaria on reflections. Q Jerus. 2016;67:86–96.
Jarman RL. Palestine and Transjordan Administration Reports 1948 – 1918. In. Edited by Jarman RL, vol. 13. London.; 1995: 171–175.
Farid MA. Ineffectiveness of DDT residual spraying in stopping malaria transmission in the Jordan Valley. Bull World Health Organ. 1954;11(4–5):765–83.
Alexander A. Fatalism and malaria elimination: a historical perspective from Palestine 100 years ago. Malariaworld J. 2018;9:4.
Garcia LS. Procedures for detecting blood parasites. In: Diagnostic Medical Parasitology. 5th edn. Edited by, editor LSS. Washington. D. C.: ASM Press; 2007.
Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pinheiro L, do Rosario VE, Thaithong S, Brown KN. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61(2):315–20.
Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419(6906):498–511.
MedicinesforMalariaVenture&WorldHealthOrganization: Methods and techniques for clinical trials on antimalarial drug efficacy: genotyping to identify parasite populations : informal consultation organized by the Medicines for Malaria Venture and cosponsored by the World Health Organization, 29–31. May 2007. In. Edited by WorldHealthOrganization. Amsterdam, The Netherlands; 2008.
Dijkman PM, Marzluf T, Zhang Y, Chang SS, Helm D, Lanzer M, Bujard H, Kudryashev M. Structure of the merozoite surface protein 1 from Plasmodium falciparum. Sci Adv. 2021;7(23).
Guimaraes LO, Wunderlich G, Alves JM, Bueno MG, Rohe F, Catao-Dias JL, Neves A, Malafronte RS, Curado I, Domingues W, et al. Merozoite surface protein-1 genetic diversity in Plasmodium malariae and Plasmodium brasilianum from Brazil. BMC Infect Dis. 2015;15:529.
Sneath PHAaS RR. Numerical Taxonomy: the Principles and Practice of Numerical classification. San Francisco: WF Freeman & Co.; 1973. p. 573.
Felsenstein J. Confidence limits on phylogenies: an Approach using the bootstrap. Evolution. 1985;39(4):783–91.
Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101(30):11030–5.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018;35(6):1547–9.
WorldHealthOrganization. : Palestine Health Profile 2015. In.; 2015.
Abdeen ZA, Sawalha SS, Eisenberger CL, Khanfar HM, Greenblatt CL, Yousef O, Schnur LF, Azmi K, Warburg A, Bader KA, et al. Epidemiology of visceral leishmaniasis in the Jenin District, West Bank: 1989–1998. Am J Trop Med Hyg. 2002;66(4):329–33.
Sawalha SS, Shtayeh MS, Khanfar HM, Warburg A, Abdeen ZA. Phlebotomine sand flies (Diptera: Psychodidae) of the palestinian West Bank: potential vectors of leishmaniasis. J Med Entomol. 2003;40(3):321–8.
Al-Jawabreh A, Ereqat S, Dumaidi K, Nasereddin A, Sawalha S, Al-Jawabreh H, Al-Jawabreh A. Case Report: Autochthonous Case of Human visceral leishmaniasis in the West Bank, Palestine. Am J Trop Med Hyg. 2020;103(4):1490–2.
Russell RC. Survival of insects in the wheel bays of a Boeing 747B aircraft on flights between tropical and temperate airports. Bull World Health Organ. 1987;65(5):659–62.
Russell RC, Paton R. In-flight disinsection as an efficacious procedure for preventing international transport of insects of public health importance. Bull World Health Organ. 1989;67(5):543–7.
Van den Ende JLL, Elsen P, Colebunders R, Demey H, Depraetere K, De Schrijver K, Peetermans WE, de Pereira P, Vogelaers D. A cluster of airport malaria in Belgium in 1995. Acta Clin Belg. 1998;53(4):259–63.
Siala E, Gamara D, Kallel K, Daaboub J, Zouiten F, Houze S, Bouratbine A, Aoun K. Airport malaria: report of four cases in Tunisia. Malar J. 2015;14:42.
Guillet P, Germain MC, Giacomini T, Chandre F, Akogbeto M, Faye O, Kone A, Manga L, Mouchet J. Origin and prevention of airport malaria in France. Trop Med Int Health. 1998;3(9):700–5.
Gallien S, Taieb F, Hamane S, De Castro N, Molina JM. Autochthonous falciparum malaria possibly transmitted by luggage-carried vector in Paris, France, February 2013. Euro Surveill 2013, 18(40).
Krajden SPD, Tobe B, Yang J, Keystone JS. Prolonged infection with Plasmodium falciparum in a semi-immune patient. Trans R Soc Trop Med Hyg. 1991;85(6):731–2.
Velasco E, Gomez-Barroso D, Varela C, Diaz O, Cano R. Non-imported malaria in non-endemic countries: a review of cases in Spain. Malar J. 2017;16(1):260.
PalestinianMinistryofHealth(PMOH). : Health Annual Report 1990–2021. In: Health Annual Report. Edited by (PHIC) PHIC. Ramallah-Palestine.; 2022.
WorldHealthOrganization. : International Health Regulations (2005), vol. 3rd; 2016.
Acknowledgements
We are thankful to the patient for participating in the case report.
Funding
This is a self-funded research study.
Author information
Authors and Affiliations
Contributions
AmJ and AN designed the study, performed data analysis, and manuscript drafting, AnJ collected the blood samples and clinical data from the patient. AN and SE performed NGS work and phylogenetic tree construction, HJ evaluated blood films from stained blood film slides.AhJ clinically evaluated the patient. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study was approved by the Al-Quds University Research Ethics Committee (Ref No. 247/REC/2022).
Consent for publication
The patient gave his written informed consent to publish the case report with all accompanying data and images.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Al-Jawabreh, A., Ereqat, S., Al-Jawabreh, A. et al. Tracking the geographical origin of Plasmodium falciparum causing a rare severe case of malaria imported into Palestine, a zero-indigenous case area. BMC Infect Dis 23, 607 (2023). https://doi.org/10.1186/s12879-023-08583-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-023-08583-4