Abstract
The epidemiological characteristics of New Delhi Metallo-β-Lactamase-Producing (NDM) Enterobacteriaceae were analyzed to provide theoretical support for clarifying the distribution characteristics of carbapenem-resistant Enterobacteriaceae (CRE) in the hospital environment and early identification of susceptible patients. From January 2017 to December 2021,42 strains of NDM-producing Enterobacteriaceae were gathered from the Fourth Hospital of Hebei Medical University, primarily Escherichia coli, Klebsiella pneumoniae, and Enterobacter cloacae. The micro broth dilution method combined with the Kirby-Bauer method was used to determine the minimal inhibitory concentrations (MICs) of antibiotics. The carbapenem phenotype was detected by the modified carbapenem inactivation method (mCIM) and EDTA carbapenem inactivation method (eCIM). Carbapenem genotypes were detected by colloidal gold immunochromatography and real-time fluorescence PCR. The results of antimicrobial susceptibility testing showed that all NDM-producing Enterobacteriaceae were multiple antibiotic resistant, but the sensitivity rate to amikacin was high. Invasive surgery prior to culture, the use of excessive amounts of different antibiotics, the use of glucocorticoids, and ICU hospitalization were clinical characteristics of NDM-producing Enterobacteriaceae infection. Molecular typing of NDM-producing Escherichia coli and Klebsiella pneumoniae was carried out by Multilocus Sequence Typing (MLST), and the phylogenetic trees were constructed. Eight sequence types (STs) and two NDM variants were detected in 11 strains of Klebsiella pneumoniae, primarily ST17, and NDM-1. A total of 8 STs and 4 NDM variants were detected in 16 strains of Escherichia coli, mainly ST410, ST167, and NDM-5. For high-risk patients who have CRE infection, CRE screening should be done as soon as feasible to adopt prompt and efficient intervention measures to prevent outbreaks in the hospital.
Similar content being viewed by others
Introduction
With the widespread use of carbapenems in clinical practice, carbapenem-resistant Enterobacteriaceae (CRE) is increasing [1]. In recent years, CRE has been reported in different countries and regions. Numerous investigations have demonstrated that the majority of CRE strains produce carbapenemases, the most prevalent carbapenemase genes were KPC and NDM [2,3,4,5]. Since the discovery of blaNDM−1 in Escherichia coli isolated from India in 2008, NDM-producing Enterobacteriaceae have been widely reported throughout the world, and Asia is thought to be the primary epidemic location. Carbapenem-resistant Klebsiella pneumoniae (CRKP) and carbapenem-resistant Escherichia coli (CREC) are the main types of Enterobacteriaceae [4,5,6].
The resistance rates of Enterobacteriaceae to imipenem and meropenem in China have climbed year over year and have now increased to nearly 10%, according to data from the China Antimicrobial Surveillance Network (CHINET). In this study, multiple methods were used to detect carbapenemases, and NDM-producing Enterobacteriaceae were screened out in 5 years. Through retrospective analysis, the distribution characteristics of NDM-type carbapenemases in the Fourth Hospital of Hebei Medical University were clarified, to monitor the development trend of carbapenemases, grasp its epidemic status, and promote the smooth implementation of hospital infection control.
Methods
Strain source
This study screened hospitalized patients in the Fourth Hospital of Hebei Medical University from January 2017 to December 2021. A total of 12,373 patients with bacterial infections were collected, 8725 patients were infected with Gram-negative bacteria, and 4987 patients were infected with Enterobacteriaceae. CRE infections (130) accounted for 1.05% of bacterial infections, and 42 CRE strains were NDM-producing Enterobacteriaceae.
Strain identification
Strain identification of 42 strains of NDM-producing Enterobacteriaceae using matrix-assisted laser desorption time-of-flight mass spectrometry (bioMérieux, France). Calibration strain: Escherichia coli ATCC8739. Quality control strain: Enterobacter aerogenes ATCC1915.
Antimicrobial susceptibility testing
The antimicrobial susceptibility testing was performed on the experimental strains using an automated VITEK®2 Compact system (bioMérieux, France) with Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC27853 as the quality control strains, and the results were determined according to the CLSI M100-S32 standard of 2022.
Carbapenemase phenotypic and genotypic assays
The carbapenemase phenotype was detected using the modified carbapenem inactivation method (mCIM) in conjunction with the EDTA carbapenem inactivation method (eCIM); the genotypes of the carbapenemase were determined using the colloidal gold immunochromatography kit (Changsha Zhongjie Biotechnology Co., Ltd.) and real-time fluorescence PCR kit (Cepheid, USA), and the consistency of the two detection methods was verified.
Information collection from medical records
The medical records of patients infected with NDM-producing Enterobacteriaceae were checked, and the following observations were recorded and counted: patients’ hospitalization department, gender, age, underlying disease, invasive operation before culture, history of antibiotics use, and hospitalization time.
MLST and evolutionary analysis
The whole genome shotgun (WGS) strategy was used to sequence 11 strains of Klebsiella pneumoniae and 16 strains of Escherichia coli based on the Illumina NovaSeq sequencing platform to obtain bacterial genome sequences; MLST 2.0 (https://cge.cbs.dtu.dk/services/MLST/ ) for multilocus sequence typing, detecting the STs of the strains and obtaining the housekeeping gene sequences of the strains; using MEGA 11 (https://www.megasoftware.net) software for multiple sequence alignment and phylogenetic tree construction, and using iTOL online beautification tool (https://itol.embl.de) for Visualization.
Accession number
The complete genome sequences of Klebsiella pneumoniae and Escherichia coli are deposited in the GenBank database as the accession number PRJNA903849 and PRJNA903692.
Results
Strain distribution
Forty-two strains of NDM-producing Enterobacteriaceae in total, including Escherichia coli (16, 38.1%), Klebsiella pneumoniae (11, 26.2%), Enterobacter cloacae (8, 19.1%), Citropebacterium freundii (3, 7.1%), Klebsiella oxytoca (3, 7.1%), and Enterobacter aerogenes (1, 2.4%), were gathered for this research. Figure 1 displays the annual changes in the proportion of NDM-producing Enterobacteriaceae in our hospital since 2017.
Antimicrobial susceptibility testing
The antimicrobial susceptibility testing results of the experimental strains are shown in Table 1. All NDM-producing Enterobacteriaceae showed multiple antibiotic resistance, highly resistant to meropenem and imipenem, with very low sensitivity to cephalosporins and penicillins and high sensitivity to amikacin, among which the sensitivity rates of Klebsiella pneumoniae and Enterobacter cloacae reached 100%.
Antibiotics-resistant phenotypic screening and genotype testing
It was verified that all 42 strains collected in this study were metallo-carbapenemase-producing Enterobacteriaceae; the results of CRE genotype detection by colloidal gold immunochromatography and real-time fluorescence PCR showed high consistency, and the antibiotics-resistant genotypes of all 42 CRE strains were NDM.
Bioinformatics analysis
Eleven strains of NDM-producing Klebsiella pneumoniae and 16 strains of NDM-producing Escherichia coli were analyzed by the ResFinder database (Additional file 2). It was found that all strains had multiple antibiotic resistance genes, and all carried carbapenem resistance genes. The majority of strains carried extended-spectrum β-lactamases (blaCTX, blaSHV, blaTEM, etc.), and other antibiotic resistance genes such as sulfonamides (sul1, sul2, sul3, etc.) and aminoglycosides (aac(6’)-Ib-cr, aph(3’)-Ia, etc.).
Clinical characteristics of patients infected with CRE
There were 42 patients altogether, 28 of whom were men. 78.6% of those infected with CRE were over the age of 50, 35.7% had undergone surgery within one month. More than half of the patients had malignant tumors (23/42, 54.8%), and most of them had combined underlying diseases such as cardiovascular and cerebrovascular diseases, pulmonary diseases, hypertension, and a history of smoking. More than 50% of infected patients had used indwelling devices such as arteriovenous catheters and drainage tubes. 57.1% of CRE-infected patients used three or more antibiotics, and most patients used antibiotics for more than 14 days. In addition, nearly half of the patients with CRE infection (20/42, 47.6%) had ICU hospitalization experience. The results are shown in Table 2.
Sequence typing and phylogenetic analysis
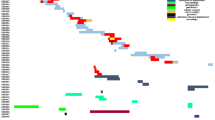
In this study, CRKP was mainly divided into three evolutionary clusters, and CREC was divided into four different evolutionary clusters, as shown in Fig. 2. Multilocus sequence typing (MLST) showed that 11 strains of Klebsiella pneumoniae had 8 STs, mainly ST17 strains, with 3 strains detected; the genotypes of NDM-producing Klebsiella pneumoniae were NDM-1 and NDM-5, of which 10 strains were NDM-1. A total of 8 STs were found in 16 Escherichia coli strains, with ST410 and ST167 strains having the most (5 strains each); NDM-producing Escherichia coli genotypes comprised NDM-4, NDM-5, NDM-7, and NDM-9, with NDM-5 being the most common.
Phylogenetic analysis of NDM-producing Klebsiella pneumoniae and Escherichia coli.Annotated information includes the specimen type, ward, MLST typing, CRE genotype, and the year of the strains a. Phylogenetic analysis of NDM producing Klebsiella pneumoniae b. Phylogenetic analysis of NDM-producing Escherichia coli
Discussion
Data from the China Antimicrobial Surveillance Network showed that among the metallocarbapenemases (MBLs) clinically isolated from general hospitals in China in 2020, the NDM type was the most found, followed by the IMP type, and the VIM type was the least discovered. In this investigation, we discovered that 42 instances of metallocarbapenemase-producing Enterobacteriaceae collected in our hospital from January 2017 to December 2021 were of the NDM type, which is comparable with the prevalent status in China. We also discovered that since 2017, the prevalence of NDM carbapenemases has progressively increased in our hospital, and the range of strains has extended, similar to those reported in other Asian countries [7,8,9]. Compared with other carbapenemases, blaNDM transmission has broader epidemiological characteristics, including (1) a wide range of Gram-negative hosts; (2) Escherichia coli and Klebsiella pneumoniae are used as conditional pathogens in the intestinal flora and can lead to patient infection through flora displacement; (3) the high population density in Southeast Asia, where it is widespread, makes it easy to disseminate antibiotics-resistant genes; (4) other resistance genes are often present on plasmids carrying blaNDM [10].
According to the results of the antimicrobial susceptibility testing, Enterobacteriaceae carrying blaNDM gene were highly resistant to most β-lactams and were sensitive to quinolones and aminoglycosides, especially amikacin. The vast majority of NDM-producing Enterobacteriaceae were sensitive to amikacin. This is consistent with the results of the above bioinformatics analysis, indicating that the coexistence of multiple antibiotic resistance genes gives the host bacteria multiple antibiotic resistance. It has been shown that the antibiotic resistance of clinical isolates to cephalosporins and aminoglycosides in developing countries, including China, is higher than that in developed countries [11]. Therefore, clinical attention should be paid to standardizing the principles and systems for the use of antimicrobial agents, and scientifically and rationally adjusting the dosing regimen.
In the present study, most of the patients with CRE infection were male, and the majority of elderly males were over 50 years old, accounting for 59.5%. As shown in Table 2, elderly patients, hypertension, diabetes, ICU stay, pulmonary disease, and various invasive operations were important clinical characteristics for CRE infection, which is generally consistent with the previous study by Pang F et al. [12]. The results of this study showed that 83% of patients received antibiotic therapy within 30 days prior to a positive culture, and more than half of these patients had a history of receiving more than three or using more than 14 days of antibiotics, 25 (25/42, 59.5%) patients received 3rd or 4th generation cephalosporins, and 17 (17/42, 40.5%) patients received carbapenems or penicillins. Overexposure to multiple antibiotics and a history of 3rd or 4th generation cephalosporins and carbapenems prior to infection explain the high rate of CRE infections [13, 14].
MLST analysis revealed the genetic diversity of CRE strains. It was shown that NDM-producing Klebsiella pneumoniae are distributed among numerous STs, and ST11, ST14, ST15, and ST147 are relatively common types that have been identified in several countries [15], although the current evidence is insufficient to prove that ST11, ST14, ST15, and ST147 are the prevalent clonal lineages mediating the spread of blaNDM internationally, their distribution in several countries warrants further study for their distribution. The 11 NDM-producing Klebsiella pneumoniae strains in this study include 8 STs,2 STs (ST11 and ST14) are consistent with it, and the other six are less common and have some local specificity. Twenty-four variants of NDM have been identified so far, among which NDM-1 has the broadest host spectrum, with Klebsiella pneumoniae and Escherichia coli being the main carriers of blaNDM−1 [16], and NDM-1 was likewise the major NDM variant of Klebsiella pneumoniae in this study.
The 16 NDM-producing Escherichia coli strains in this study included 8 STs, mainly ST410 and ST167, with 5 strains detected each. Compared with other ST strains, the affinity between ST167 and ST617 is very close, with ST167 strains being the high-risk epidemic type in China [17, 18], and four of the five ST167 strains were NDM-5 variants, suggesting that ST167 is closely related to blaNDM−5 to some extent, which is consistent with previous studies [19, 20]. Additionally, even though 5 ST167 strain cases were found in this study, they were all from different wards and different years, preventing nosocomial infection outbreaks. In contrast, 2 ST410 strain cases were found in the nephrology department in June 2020 and were both NDM-5 variants, suggesting the possibility of nosocomial local transmission of CREC. NDM-5 was discovered in 2011 in a strain of Escherichia coli in the United Kingdom [21], and it differs from NDM-1 by having higher carbapenemase activity and amino acid changes at positions 88 (ValLeu) and 154 (MetLeu). Since then, strains carrying blaNDM−5 have been reported in numerous countries including Singapore, India, Japan, China, Australia, and the United States, and NDM-5 has become the second most reported NDM variant after NDM-1 in the world [22,23,24]. Notably, in previous reports, NDM-1 was likewise the most prevalent NDM variant among CRE strains in China, but our study showed that NDM-5 was predominant among CREC strains and accounted for 81.25%, and a similar situation was found in a teaching hospital in Chongqing [20], which reveals that CREC carrying blaNDM−5 is gradually becoming a new potential threat, and there is an urgent need for clinical attention and effective interventions to control its further spread.
Conclusion
In conclusion, we should expand research on the molecular epidemiology of CRE and the prevalence trend of New Delhi Metallo-β-lactamase-producing Enterobacteriaceae to offer a scientific foundation for early detection of patients with high-risk CRE infection, early screening of patients with high-risk CRE infection, and prompt and effective implementation of therapies to improve patients’ clinical outcomes.
Data Availability
The datasets that support the findings of this study are available in the NCBI GenBank database, https://www.ncbi.nlm.nih.gov/bioproject/. Accession codes are PRJNA903849 and PRJNA903692.
References
Lutgring JD. Carbapenem-resistant Enterobacteriaceae: an emerging bacterial threat. Semin Diagn Pathol. 2019;36(3):182–6.
Alotaibi F. Carbapenem-Resistant Enterobacteriaceae: an update narrative review from Saudi Arabia. J Infect Public Health. 2019;12(4):465–71.
Takeuchi D, Kerdsin A, Akeda Y, Sugawara Y, Sakamoto N, Matsumoto Y, Motooka D, Ishihara T, Nishi I, Laolerd W et al. Nationwide surveillance in Thailand revealed genotype-dependent dissemination of carbapenem-resistant Enterobacterales.Microb Genom2022, 8(4).
Wang Q, Wang X, Wang J, Ouyang P, Jin C, Wang R, Zhang Y, Jin L, Chen H, Wang Z, et al. Phenotypic and genotypic characterization of Carbapenem-resistant Enterobacteriaceae: data from a longitudinal large-scale CRE Study in China (2012–2016). Clin Infect Dis. 2018;67(suppl2):196–s205.
Zhao S, Qiu X, Tan Y, Xu M, Xu X, Ge Y, Cao H, Li M, Lin Y, Zhang Z, et al. Genomic analysis and molecular characteristics in Carbapenem-Resistant Klebsiella pneumoniae strains. Curr Microbiol. 2022;79(12):391.
Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53(12):5046–54.
Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10(9):597–602.
Wu W, Feng Y, Tang G, Qiao F, McNally A, Zong Z. NDM Metallo-β-Lactamases and Their Bacterial Producers in Health Care Settings.Clin Microbiol Rev2019, 32(2).
Han R, Shi Q, Wu S, Yin D, Peng M, Dong D, Zheng Y, Guo Y, Zhang R, Hu F. EnterobacteriaceaeDissemination of Carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) among carbapenem-resistant isolated from adult and children patients in China. Front Cell Infect Microbiol. 2020;10:314.
Wailan AM, Paterson DL. The spread and acquisition of NDM-1: a multifactorial problem. Expert Rev Anti Infect Ther. 2014;12(1):91–115.
Nasser M, Palwe S, Bhargava RN, Feuilloley MGJ, Kharat AS. Retrospective Analysis on Antimicrobial Resistance Trends and Prevalence of β-lactamases in Escherichia coli and ESKAPE Pathogens Isolated from Arabian Patients during 2000–2020.Microorganisms2020, 8(10).
Pang F, Jia X, Zhao Q, Zhang Y. Factors associated to prevalence and treatment of carbapenem-resistant Enterobacteriaceae infections: a seven years retrospective study in three tertiary care hospitals. Ann Clin Microbiol Antimicrob. 2018;17(1):13.
Wang Q, Zhang Y, Yao X, Xian H, Liu Y, Li H, Chen H, Wang X, Wang R, Zhao C, et al. Risk factors and clinical outcomes for carbapenem-resistant Enterobacteriaceae nosocomial infections. Eur J Clin Microbiol Infect Dis. 2016;35(10):1679–89.
Turner P, Pol S, Soeng S, Sar P, Neou L, Chea P, Day NP, Cooper BS, Turner C. High prevalence of antimicrobial-resistant Gram-negative colonization in hospitalized cambodian Infants. Pediatr Infect Dis J. 2016;35(8):856–61.
Yang K, Liu S, Li H, Du N, Yao J, He Q, Du Y. NDM-1-Positive K. pneumoniae at a Teaching Hospital in Southwestern China: Clinical Characteristics, Antimicrobial Resistance, Molecular Characterization, Biofilm Assay, and Virulence. Can J Infect Dis Med Microbiol 2020, 2020:9091360.
Kazmierczak KM, Rabine S, Hackel M, McLaughlin RE, Biedenbach DJ, Bouchillon SK, Sahm DF, Bradford PA. Multiyear, multinational survey of the incidence and global distribution of Metallo-β-Lactamase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2016;60(2):1067–78.
Tuem KB, Gebre AK, Atey TM, Bitew H, Yimer EM, Berhe DF. Drug Resistance Patterns of Escherichia coli in Ethiopia: A Meta-Analysis. Biomed Res Int 2018, 2018:4536905.
Garg A, Garg J, Kumar S, Bhattacharya A, Agarwal S, Upadhyay GC. Molecular epidemiology & therapeutic options of carbapenem-resistant Gram-negative bacteria. Indian J Med Res. 2019;149(2):285–9.
Huang Y, Yu X, Xie M, Wang X, Liao K, Xue W, Chan EW, Zhang R, Chen S. Widespread dissemination of Carbapenem-Resistant Escherichia coli sequence type 167 strains harboring blaNDM-5 in clinical settings in China. Antimicrob Agents Chemother. 2016;60(7):4364–8.
Zou H, Jia X, Liu H, Li S, Wu X, Huang S. Emergence of NDM-5-Producing Escherichia coli in a Teaching Hospital in Chongqing, China: IncF-Type plasmids may contribute to the prevalence of bla (NDM-) (5). Front Microbiol. 2020;11:334.
Hornsey M, Phee L, Wareham DW. A novel variant, NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agents Chemother. 2011;55(12):5952–4.
Markovska R, Stoeva T, Boyanova L, Stankova P, Schneider I, Keuleyan E, Mihova K, Murdjeva M, Sredkova M, Lesseva M, et al. Multicentre investigation of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in bulgarian hospitals - interregional spread of ST11 NDM-1-producing K. pneumoniae. Infect Genet Evol. 2019;69:61–7.
Yin C, Yang W, Lv Y, Zhao P, Wang J. Clonal spread of carbapenemase-producing Enterobacteriaceae in a region, China. BMC Microbiol. 2022;22(1):81.
van Duin D, Doi Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence. 2017;8(4):460–9.
Acknowledgements
Not applicable.
Funding
This research was funded by Hebei Provincial Department of Finance government-funded specialist capacity-building project (Grant Number 361006) and Youth Science and Technology Project of Health Department of Hebei Province (Grant Number 20221308).
Author information
Authors and Affiliations
Contributions
MSZ, JH, and JYZ designed the research; MSZ and JH performed the research; All authors analyzed the data and wrote and revise the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
The study was approved by Ethics Committee of the Fourth Hospital of Hebei Medical University, all methods were carried out in accordance with relevant guidelines and regulations. All experimental protocols were approved by the licensing committee. The study obtained the informed consent of all subjects and/or their legal guardians. There are no ethical conflicts with all the data in this article.
Consent for publication
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhao, M., He, J., Zhang, R. et al. Epidemiological characteristics of New Delhi Metallo-β-Lactamase-producing Enterobacteriaceae in the Fourth hospital of Hebei Medical University. BMC Infect Dis 23, 298 (2023). https://doi.org/10.1186/s12879-023-08242-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-023-08242-8