Abstract
Background
Identifying the biological subclasses of septic shock might provide specific targeted therapies for the treatment and prognosis of septic shock. It might be possible to find biological markers for the early prediction of septic shock prognosis.
Methods
The data were obtained from the Gene Expression Omnibus databases (GEO) in NCBI. GO enrichment and KEGG pathway analyses were performed to investigate the functional annotation of up- and downregulated DEGs. ROC curves were drawn, and their areas under the curves (AUCs) were determined to evaluate the predictive value of the key genes.
Results
117 DEGs were obtained, including 36 up- and 81 downregulated DEGs. The AUC for the MME gene was 0.879, as a key gene with the most obvious upregulation in septic shock. The AUC for the THBS1 gene was 0.889, as a key downregulated gene with the most obvious downregulation in septic shock.
Conclusions
The upregulation of MME via the renin-angiotensin system pathway and the downregulation of THBS1 through the PI3K–Akt signaling pathway might have implications for the early prediction of prognosis of septic shock in patients with pneumopathies.
Similar content being viewed by others
Background
Septic shock, characterized by circulatory and cellular abnormalities, is associated with substantial morbidity and mortality [1]. The worldwide mortality rate of septic shock was > 40% in 2016 [2, 3], making septic shock a major healthcare problem globally. It plays an important role in the morbidity and mortality of patients in the intensive care unit and results in substantial health care costs [4, 5].
Although much progress has been made in diagnosing and treating septic shock, mortality remains unacceptably high [6]. A consistent increase in sepsis has been shown over the past two decades [7], its incidence is predicted to increase rapidly because of the sepsis observed in COVID-19 [8]. It is known that certain genes and signaling pathways participate in the occurrence of septic shock in children [9]. Identifying biological subclasses of septic shock might provide specific targeted therapies for the treatment and prognosis of septic shock [10, 11]. Studying the molecular mechanism of septic shock is certainly important [9].
We hypothesized that key genes are involved in the early stage of septic shock. These genes might be involved in the initiation of septic shock, and the mediated molecular changes might affect the prognosis of the disease. Therefore, it could be possible to detect the changes in gene expression in the early stage of septic shock through gene chips. By correlating these changes with the prognosis of the disease, it might be possible to identify biological markers for the early prediction of the prognosis of septic shock.
Materials and methods
Data collection
The data was obtained from the Gene Expression Omnibus databases (GEO) in NCBI, a public functional genomics data repository. The expression dataset GSE33118 (https://www.ncbi.nlm.nih.gov/geo/; GSE33118) was obtained from the Affymetrix GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array), which was submitted by Raffelsberger et al. Septic shock by pneumopathy was studied prospectively in 20 patients, whose blood samples were taken within 12 h of diagnosis. Preparation, processing, and analysis of data were performed using the R software (version 3.6.3). The flowchart of this study is shown in Fig. 1.
Identification of differentially expressed genes
The limma package in R was used to normalize and correct the data [12]. The expression data were screened for differentially expressed genes (DEGs) between different outcomes with the criteria of |log2fold change (FC)|> 1 and adjusted P-value < 0.05 [13]. Then, the identified DEGs were divided into upregulated and downregulated DEGs.
DEGs analyze via GO and KEGG
Gene Ontology (GO) [14] enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) [15,16,17] pathway analysis were performed to investigate the functional annotation of the up- and downregulated DEGs. The expression matrix of the up- and downregulated DEGs were analyzed by GO and KEGG enrichment to determine whether they show statistically significant differences between different outcomes. A P-value < 0.05 was considered statistically significant.
ROC analysis of the significantly enriched DEGs
Up- and downregulated DEGs were considered candidate genes to predict the outcome. Receiver operating characteristic (ROC) curves were drawn. Their areas under the curve (AUCs) were determined to evaluate the predicted value of the key genes using the pROC package in R [18].
Results
Identification of DEGs
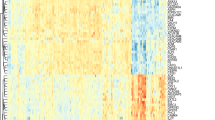
There were 54,613 genes in the GSE33118 dataset, including 29,647 upregulated and 24,939 downregulated genes. Finally, 117 DEGs were identified, including 36 up- and 81 downregulated DEGs. The volcano map of all DEGs is displayed in Fig. 2A. The heatmaps of the top 36 upregulated genes and the top 81 downregulated genes are displayed in Fig. 2B, C.
GO enrichment and KEGG pathway of DEGs
GO categories and KEGG pathways were used to evaluate the up- and downregulated genes. The upregulated DEGs were significantly enriched in Molecular Function (MF), such as phosphatidylinositol-3-phosphate binding, sodium channel regulator activity, potassium channel regulator activity, extracellular matrix structural constituent conferring tensile strength, chemokine binding, chemokine receptor activity, G protein-coupled chemoattractant receptor activity, and C–C chemokine binding, as well as in Biological Process (BP), such as regulation of polysaccharide metabolic process, amyloid-beta clearance, regulation of glycogen metabolic process, and long-term memory (Fig. 3A). The KEGG pathway results revealed that the upregulated DEGs were significantly enriched in the renin-angiotensin system, aldosterone-regulated sodium reabsorption, ECM-receptor interaction, and hematopoietic cell lineage (Fig. 3B). Among those results, we will focus on the key genes, such as MME, SGK1, and COL9A3 (Fig. 3C, D).
The downregulated DEGs were significantly enriched in cellular components (CC), such as protein kinase complex, endoplasmic reticulum-Golgi intermediate compartment, serine/threonine-protein kinase complex, and cyclin-dependent protein kinase holoenzyme complex (Fig. 4A). The KEGG pathway results showed that the downregulated DEGs were significantly enriched in the PI3K–Akt signaling pathway, p53 signaling pathway, cell cycle, ECM-receptor interaction, and hematopoietic cell lineage (Fig. 4B). We will focus on the key genes, such as CD24, MS4A1, HMMR, DDIT4, TCL1A, AREG, BUB1, TTK, CCNB2, THBS1, and RRM2 (Fig. 4C, D).
Identification of the key genes associated with the outcome
There were three upregulated and 11 downregulated key genes. Among them, all the key genes showed that they could predict the outcome accurately with AUCs > 0.7, except for BUB1 (Fig. 5). The AUC for the MME gene was 0.879, as a key gene with the most obvious change in expression among the upregulated genes. The AUC for the THBS1 gene was 0.889, as a key downregulated gene with the most obvious change in expression among the downregulated genes. The prognostic cutoff concentration to predict the outcome was chosen after ROC analysis (Fig. 5).
Discussion
Sepsis is a main cause of critical illness and mortality all over the world [19, 20]. Septic shock reflects a more severe illness with a higher likelihood of death than sepsis alone [1]. Septic shock results in multiorgan dysfunction, including the liver, kidney, and lung [1]. Clinical symptoms of early septic shock are often nonspecific, so they are easy to miss. The identification of the early signs of septic shock might help a timely diagnosis and initiate therapy faster [21, 22]. This study examined the biological subclasses using bioinformatics and could provide a foundation for the molecular diagnosis and prognosis of septic shock from pneumopathies.
In this study, as a significantly upregulated gene, MME (membrane metalloendopeptidase) might play an important role in the prognosis of septic shock through the renin-angiotensin system pathway [23], which is activated to increase the arterial blood pressure in sepsis [24]. Indeed, MME can produce Ang-(1–7) (an active form of angiotensin) from Ang-(1–9) [23]. Ang-(1–7) is a potent vasodilator that might be participating in the dramatic drop in blood pressure often observed in septic shock [25]. Still, Ang-(1–7) appears to have protective actions in sepsis [26]. On the other hand, MME has also been involved in Ang-(1–12) metabolism, which is known to activate the renin-angiotensin system [23]. Excessive activation of the renin-angiotensin system might further worsen the outcomes of sepsis [27]. Fernandes et al. revealed that a partial blockade of the renin-angiotensin system might provide an opportunity to improve the outcomes of sepsis-induced refractoriness to vasoconstrictors [28]. Furthermore, the renin-angiotensin system has been recognized to play a major role in some biological processes such as coagulation, apoptosis, and inflammation [29, 30]. Still, the exact role of MME in sepsis remains to be determined.
In this study, as a significantly downregulated gene, THBS1 might play an important part in the prognosis of septic shock through the PI3K–Akt signaling pathway. Mizuta et al. [31] suggested that activating the PI3K/Akt signaling pathway contributes to suppressing endothelial apoptosis, inhibits lung hemorrhage and edema, and improves murine survival. The morbidity and mortality caused by myocardial infarction might be decreased via the stimulation of the PI3K/Akt-dependent cascade [32]. It was revealed that suppressing inflammatory and antioxidant responses through the PI3K/Akt pathway might mitigate the effects of sepsis [33]. In this study, besides THBS1, DDIT4, TCL1A, and AREG might be biological markers in the prognosis of septic shock by acting via PI3K/Akt signaling pathway. Still, the role of THBS1 in the regulation of the PI3K–Akt signaling pathway in cancer is relatively well defined [34], but its role in sepsis remains to be determined.
Different metabolomic patterns might have a major impact on the development of novel diagnostic methods for the early diagnosis and prognosis of septic shock [35]. The heterogeneity between DEGs and the specific signaling pathways prompted us to select one gene to be used as a prognostic marker of the outcome. In this study, prognostic cutoff concentrations for predicting the outcome were selected using a ROC curve analysis.
The potential limitations of our study should be considered. The datasets we used were obtained from a public database. This dataset only included data about septic shock from pneumopathies. All results were obtained using bioinformatics. Future studies are currently being designed for examining the exact roles of MME and THBS1 in the renin-angiotensin system and PI3K/Akt signaling pathways in septic shock from pneumopathies. Further experiments are needed to verify the results of this study and expand the generalizability to septic shock from other sources.
Conclusion
In summary, the upregulation of MME and its role in the renin-angiotensin system pathway and the downregulation of THBS1 and its role in the PI3K–Akt signaling pathway might have important implications for the early diagnosis and prognosis of septic shock from pneumopathies. This study suggests that the molecular typing of septic shock could reveal diagnostic, prognostic, and therapeutic biomarkers for patients with septic shock, allowing early diagnosis and management. Still, the biomarkers need to be refined, and prognosis models need to be built.
Availability of data and materials
The datasets analyzed during the current study are publicly available in the [Gene Expression Omnibus databased] repository/database, [https://www.ncbi.nlm.nih.gov/geo/; GSE33118].
References
Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche JD, Coopersmith CM, et al. The third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA. 2016;315(8):801–10.
Rhodes A, Evans LE, Alhazzani W, Levy MM, Antonelli M, Ferrer R, Kumar A, Sevransky JE, Sprung CL, Nunnally ME, et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock: 2016. Intensive Care Med. 2017;43(3):304–77.
Martin GS, Mannino DM, Eaton S, Moss M. The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med. 2003;348(16):1546–54.
Fleischmann-Struzek C, Mikolajetz A, Schwarzkopf D, Cohen J, Hartog CS, Pletz M, Gastmeier P, Reinhart K. Challenges in assessing the burden of sepsis and understanding the inequalities of sepsis outcomes between National Health Systems: secular trends in sepsis and infection incidence and mortality in Germany. Intensive Care Med. 2018;44(11):1826–35.
Zhang J, Luo Y, Wang X, Zhu J, Li Q, Feng J, He D, Zhong Z, Zheng X, Lu J, et al. Global transcriptional regulation of STAT3- and MYC-mediated sepsis-induced ARDS. Ther Adv Respir Dis. 2019;13:1753466619879840.
Rudd KE, Johnson SC, Agesa KM, Shackelford KA, Tsoi D, Kievlan DR, Colombara DV, Ikuta KS, Kissoon N, Finfer S, et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: analysis for the Global Burden of Disease Study. Lancet. 2020;395(10219):200–11.
Lee CC, Yo CH, Lee MG, Tsai KC, Lee SH, Chen YS, Lee WC, Hsu TC, Lee SH, Chang SS. Adult sepsis—a nationwide study of trends and outcomes in a population of 23 million people. J Infect. 2017;75(5):409–19.
Lin L, Lu L, Cao W, Li T. Hypothesis for potential pathogenesis of SARS-CoV-2 infection—a review of immune changes in patients with viral pneumonia. Emerg Microbes Infect. 2020;9(1):727–32.
Yang J, Zhang S, Zhang J, Dong J, Wu J, Zhang L, Guo P, Tang S, Zhao Z, Wang H, et al. Identification of key genes and pathways using bioinformatics analysis in septic shock children. Infect Drug Resist. 2018;11:1163–74.
Wong HR, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Willson DF, Freishtat RJ, Anas N, Meyer K, Checchia PA, et al. Identification of pediatric septic shock subclasses based on genome-wide expression profiling. BMC Med. 2009;7:34.
Wynn JL, Cvijanovich NZ, Allen GL, Thomas NJ, Freishtat RJ, Anas N, Meyer K, Checchia PA, Lin R, Shanley TP, et al. The influence of developmental age on the early transcriptomic response of children with septic shock. Mol Med. 2011;17(11–12):1146–56.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47.
Liu J, Nie S, Gao M, Jiang Y, Wan Y, Ma X, Zhou S, Cheng W. Identification of EPHX2 and RMI2 as two novel key genes in cervical squamous cell carcinoma by an integrated bioinformatic analysis. J Cell Physiol. 2019;234(11):21260–73.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9.
Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30.
Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28(11):1947–51.
Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49(D1):D545–51.
Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Muller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12:77.
Vincent JL, Marshall JC, Namendys-Silva SA, Francois B, Martin-Loeches I, Lipman J, Reinhart K, Antonelli M, Pickkers P, Njimi H, et al. Assessment of the worldwide burden of critical illness: the intensive care over nations (ICON) audit. Lancet Respir Med. 2014;2(5):380–6.
Fleischmann C, Scherag A, Adhikari NK, Hartog CS, Tsaganos T, Schlattmann P, Angus DC, Reinhart K, International Forum of Acute Care T. Assessment of global incidence and mortality of hospital-treated sepsis. Current estimates and limitations. Am J Respir Crit Care Med. 2016;193(3):259–72.
Eitze S, Fleischmann-Struzek C, Betsch C, Reinhart K, vaccination60+ study g. Determinants of sepsis knowledge: a representative survey of the elderly population in Germany. Crit Care. 2018;22(1):273.
Lu J, Li Q, Wu Z, Zhong Z, Ji P, Li H, He C, Feng J, Zhang J. Two gene set variation indexes as potential diagnostic tool for sepsis. Am J Transl Res. 2020;12(6):2749–59.
Nehme A, Zouein FA, Zayeri ZD, Zibara K. An update on the tissue renin angiotensin system and its role in physiology and pathology. J Cardiovasc Dev Dis. 2019;6(2):14.
Doerschug KC, Delsing AS, Schmidt GA, Ashare A. Renin-angiotensin system activation correlates with microvascular dysfunction in a prospective cohort study of clinical sepsis. Crit Care. 2010;14(1):R24.
Padda RS, Shi Y, Lo CS, Zhang SL, Chan JS. Angiotensin-(1–7): a novel peptide to treat hypertension and nephropathy in diabetes? J Diabetes Metab. 2015. https://doi.org/10.4172/2155-6156.1000615.
Tsai HJ, Shih CC, Chang KY, Liao MH, Liaw WJ, Wu CC, Tsao CM. Angiotensin-(1–7) treatment blocks lipopolysaccharide-induced organ damage, platelet dysfunction, and IL-6 and nitric oxide production in rats. Sci Rep. 2021;11(1):610.
Forrester SJ, Booz GW, Sigmund CD, Coffman TM, Kawai T, Rizzo V, Scalia R, Eguchi S. Angiotensin II signal transduction: an update on mechanisms of physiology and pathophysiology. Physiol Rev. 2018;98(3):1627–738.
Fernandes D, Pacheco LK, Sordi R, Scheschowitsch K, Ramos GC, Assreuy J. Angiotensin II receptor type 1 blockade improves hyporesponsiveness to vasopressors in septic shock. Eur J Pharmacol. 2021;897:173953.
Zhang L, Ren Z, Yang Q, Ding G. Csk regulates angiotensin II-induced podocyte apoptosis. Apoptosis. 2016;21(7):846–55.
Yang Y, Tian T, Wang Y, Li Z, Xing K, Tian G. SIRT6 protects vascular endothelial cells from angiotensin II-induced apoptosis and oxidative stress by promoting the activation of Nrf2/ARE signaling. Eur J Pharmacol. 2019;859:172516.
Mizuta Y, Akahoshi T, Guo J, Zhang S, Narahara S, Kawano T, Murata M, Tokuda K, Eto M, Hashizume M, et al. Exosomes from adipose tissue-derived mesenchymal stem cells ameliorate histone-induced acute lung injury by activating the PI3K/Akt pathway in endothelial cells. Stem Cell Res Ther. 2020;11(1):508.
Chen L, Liu P, Feng X, Ma C. Salidroside suppressing LPS-induced myocardial injury by inhibiting ROS-mediated PI3K/Akt/mTOR pathway in vitro and in vivo. J Cell Mol Med. 2017;21(12):3178–89.
Santos DMD, Da Silva EAP, Oliveira JYS, Marinho YYM, Santana IR, Heimfarth L, Pereira EWM, Junior LJQ, Assreuy J, Menezes IAC, et al. The therapeutic value of hydralazine in reducing inflammatory response, oxidative stress, and mortality in animal sepsis: involvement of the PI3K/AKT pathway. Shock. 2021;56(5):782–92.
Chen X, Guo ZQ, Cao D, Chen Y, Chen J. MYC-mediated upregulation of PNO1 promotes glioma tumorigenesis by activating THBS1/FAK/Akt signaling. Cell Death Dis. 2021;12(3):244.
Neugebauer S, Giamarellos-Bourboulis EJ, Pelekanou A, Marioli A, Baziaka F, Tsangaris I, Bauer M, Kiehntopf M. Metabolite profiles in sepsis: developing prognostic tools based on the type of infection. Crit Care Med. 2016;44(9):1649–62.
Acknowledgements
We want to thank the participants for providing the information used in this study and for kindly making the arrangements for data collection.
Funding
This study was supported by the Natural Science Foundation of Fujian Province (Grant No. 2020J011068), the Startup Fund for scientific research, Fujian Medical University (Grant No. 2018QH1160), the Natural Science Foundation of Fujian Province (Grant No. 2019J01175), and the Young and Middle-aged Talents Training Project of Fujian Provincial Health Commission (Grant No. 2018-ZQN-1).
Author information
Authors and Affiliations
Contributions
SS and XP performed the formal analysis, methodology, and writing-original draft. HF, and SZ performed the data curation and resources. SS and WL performed the conceptualization, formal analysis, writing-original draft, and project administration. WL is the guarantor of this manuscript and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study has been approved by the Fujian Provincial Hospital Ethics Committee. The study followed the principles of the Declaration of Helsinki. All data were obtained from a public domain database.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Shi, S., Pan, X., Feng, H. et al. Identification of transcriptomics biomarkers for the early prediction of the prognosis of septic shock from pneumopathies. BMC Infect Dis 21, 1190 (2021). https://doi.org/10.1186/s12879-021-06888-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-021-06888-w