Abstract
Background
The worldwide spread of carbapenemase-producing Enterobacteriaceae (CPE) has reduced the clinical utility of carbapenems. Plasmids often play an important role in the spread of genes encoding drug-resistance factors, especially in the horizontal transfer of these genes among species of Enterobacteriaceae. This study describes a patient infected with three species of CPE carrying an identical transferrable IncL/M plasmid.
Methods
Clinical isolates of CPE were collected at St. Luke’s International Hospital, Tokyo, Japan, from 2015 to 2019. Three species of CPE isolates, Enterobacter cloacae, Klebsiella aerogenes and Serratia marcescens, were isolated from a patient who developed severe gallstone pancreatitis associated with bloodstream infection, with all three isolates producing IMP-1 metallo-β-lactamase. The complete sequences of the plasmids of the three isolates were determined by both MiSeq and MinION. The medical chart of this patient was retrospectively reviewed conducted to obtain relevant clinical information.
Results
The three CPE species carried an IncL/M plasmid, pSL264, which was 81,133 bp in size and harbored blaIMP-1. The genetic environment surrounding blaIMP-1 consisted of int1-blaIMP-1-aac(6’)-IIc-qacL-qacEdelta1-sul1-istB-IS21. Conjugation experiments showed that S. marcescens could transmit the plasmid to E. cloacae and K. aerogenes. In contrast, pSL264 could not transfer from E. cloacae or K. aerogenes to S. marcescens.
Conclusion
The IncL/M plasmid pSL264 harboring blaIMP-1 was able to transfer among different species of Enterobacteriaceae in a patient receiving long-term antimicrobial treatment. The worldwide emergence and spread of IncL/M plasmids harboring carbapenemase-encoding genes among species of Enterobacteriaceae is becoming a serious public health hazard.
Similar content being viewed by others
Background
Carbapenems are often the treatment of last resort for patients with serious infections caused by gram-negative bacteria. The worldwide emergence of carbapenemase-producing Enterobacteriaceae (CPE) has limited the clinical utility of this class of antimicrobial agents [1]. The most common carbapenemases among CPE are Klebsiella pneumoniae carbapenemases (Amber class A), IMP-type metallo-β-lactamases (MBLs), VIM-type MBLs, NDM-type MBLs (class B), and OXA-48-like (class D) enzymes [2]. MBLs produced by gram-negative bacteria confer resistance to all β-lactams, except for aztreonam, and are characterized by their efficient hydrolysis of carbapenems [2, 3]. IMP-1 was first discovered in Japan in 1991 [4] and IMP-producing Pseudomonas aeruginosa and Enterobacteriaceae are frequently detected in patients in the Asia–Pacific region [2, 4,5,6,7].
Plasmids often play an important role in the spread of genes encoding drug resistance factors, with these plasmids often involved in the horizontal transfer of drug resistance genes among Enterobacteriaceae [8]. For example, the plasmid IncX4, which harbors mcr-1, a gene associated with colistin resistance, was shown to easily transfer between Escherichia coli and K. pneumoniae [9]. In addition, IncX3 plasmids carrying blaNDMs are effectively transferred among Enterobacteriaceae species and contribute to the worldwide dissemination of NDM producers [10, 11]. IncFII(K) plasmids carrying blaIMPs and several Inc type plasmids, including IncL/M, IncN2, IncHI1B-IncFIB (Mar) and IncX3-IncC-like plasmids, carrying blaVIMs have contributed to the global spread of blaIMPs and blaVIMs among different species of Enterobacteriaceae [12].
Few reports have described the in vitro and in vivo horizontal transfer of identical carbapenemase-encoding plasmids [13,14,15,16]. This study describes a patient infected with three species of Enterobacteriaceae, all of which carried a transferrable IncL/M plasmid producing IMP-1.
Methods
Bacterial strains and antimicrobial susceptibility
All clinical isolates of Enterobacteriaceae not susceptible to carbapenems were prospectively collected and stored at the microbiological laboratory of St. Luke’s International Hospital, Tokyo, Japan, from April 2015 to May 2019. Initial identification and susceptibility tests were performed using Microscan WalkAway 96 Plus (Beckman-Coulter) and Microscan Neg Series NC-EN2J (Beckman-Coulter). Susceptibility results were interpreted in accordance with the Performance Standards for Antimicrobial Susceptibility Testing M100-25 of the Clinical Laboratory Standards Institute (CLSI) [17]. These isolates were screened for production of carbapenemase by a modified carbapenem inactivation method. Thereafter, MBLs were detected using sodium mercaptoacetic acid. During the screening of CPE, one patient was found to harbor three species of CPE, Enterobacter cloacae, Serratia marcescens and Klebsiella aerogenes.
Identification and whole genome sequencing of CPE harboring bla IMP-1
The CPE isolates were screened for the presence of blaIMP-1-like by PCR using the primers IMP-F (5’-ATGAGCAAGTTATCTGTATTCTTTA-3’) and IMP-R (5’-TTAGTTGCTTGGTTTTGATGGTTTT-3’). All PCR products were sequenced using ABI PRISM 3500XL DNA Analyzer (Applied Biosystems, Foster, CA). The whole genome of each isolate was extracted using DNeasy Blood and Tissue kits (Qiagen, Tokyo, Japan) for MiSeq (Illumina, San Diego, CA) and QIAGEN Genomic-tip 100/G and Genomic DNA Buffer Set (Qiagen) for MinION (Oxford Nanopore Technologies, Oxford, UK). Whole genomes and plasmids of the three isolates were sequenced by MiSeq platform using 600 cycle Reagent Kit v.3 and MinION platform using R9.4 flow cell (FLO-MIN106), according to the manufacturers’ instructions. The sequence reads generated by MiSeq were quality trimmed and filtered using CLC Genomics Workbench v11 (CLC bio, Aarthus, Denmark). MinION data were base called by Guppy v3.6.1 (Oxford Nanopore), trimmed by NanoFilt v2.2 (https://github.com/wdecoster/nanofilt), and adaptors trimmed by Porechop v0.2.3 (https://github.com/rrwick/Porechop). The long reads generated by MinION and the short reads generated by Miseq were assembled and polished using Unicycler v0.4.7 [18]. The sequence of drug resistance genes and plasmid typing were determined using Resfinder 4.1 and PlasmidFinder 2.1, respectively, from the Center for Genomic Epidemiology (CGE) (https://cge.cbs.dtu.dk/services/).
Structure of the plasmids harboring bla IMP-1
The sequences of the plasmids in the three isolates were compared using GENETYX-MAC ver. 19.0.1 (GENETYX Co.), and the detailed genetic structure of the plasmid in E. cloacae was determined by the Rapid Annotation using Subsystem Technology version 2.0 (https://rast.nmpdr.org) [19]. Whole genome sequences of the plasmids were analyzed by BLAST® website (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed September 21, 2020) and in silico MolecularCloning Genomics Edition v7 (in silico biology, Inc. Kanagawa, Japan).
Conjugation of the plasmid among the isolates
The type strains of E. cloacae (NBRC13535), K. aerogenes (NBRC13534) and S. marcescens (NBRC102204) were plated on Mueller–Hinton broth (MHB) containing 50 µg/ml rifampicin. The grown cells were selected as recipient strains. The donor strains were cultivated in MHB containing ceftazidime and the recipient strains in MHB overnight. The cells were harvested by centrifugation (5000×g for 3 min), washed three times with PBS and resuspended in PBS to an OD = 3.5. Donor and recipient cells, mixed at an optimal ratio of 1:3 [20], were incubated for 3 h at 37 ºC and plated on selective media, consisting of MHB containing 50 µg/ml rifampicin or 50 µg/ml ceftazidime and 50 µg/ml rifampicin, for 18 h. The conjugation frequencies of donor cells were calculated based on the number of colonies on each selective medium.
Multilocus sequence typing and antimicrobial resistance genes
Multilocus sequence typing (MLST) was performed using MLST 2.0 (https://cge.cbs.dtu.dk/services/MLST-2.0/) [21], and antimicrobial resistant genes of E. cloacae SL264 were determined using ResFinder 4.1 (https://cge.cbs.dtu.dk/services/ResFinder/) [22].
Clinical information
The medical chart of the patient was retrospectively reviewed conducted to obtain relevant clinical information.
Nucleotide sequence accession number
The chromosome and two plasmids (pSL264 and pSL264-2) sequences of E. cloacae SL264 have been deposited at GenBank under accession number AP024913, AP024914 and AP024915, respectively. The chromosome and the plasmid (pSL267) sequences of S. marcescens SL267 were AP024916 and AP024917, respectively. The chromosome and two plasmids (pSL269 and pSL269-2) sequences of K. aerogenes SL269 have been deposited at GenBank under accession number AP024918, AP024919 and AP024920, respectively.
Results
Description of the patient
The patient was an 86-year-old man with a history of pancreatic cancer who developed severe gallstone pancreatitis and complicated pancreatic cyst infection. Of note, he was empirically treated with meropenem and a surveillance culture of his pancreatic fluid yielded an IMP-1-producing strain of E. cloacae, designated SL264, which was considered colonization (Table 1). Two months after the initial event, an IMP-1-producing strain of S. marcescens, designated SL267, was isolated from the pancreatic drainage tube (Table 1). Five days later, however, the patient developed a high fever. Culture of a blood sample resulted in the isolation of an IMP-1-producing strain of K. aerogenes, designated SL269 (Table 1). The patient was successfully treated with prolonged infusion of meropenem, colistin and tigecycline for 14 days.
Structure of the plasmid harboring bla IMP-1
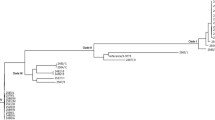
The plasmid identified in E. cloacae SL264 was designated pSL264; it was found to be 81,133 bp in length and have a genetic structure of int1-blaIMP-1-aac(6’)-IIc-qacL-qacEdelta1-sul1-istB-IS21 (Fig. 1). The blaIMP-1 gene was on a class 1 integron with a unique structure. The sequence of qacEdelta1-sul1-istB-IS21 was 99.25% identical to that of the plasmid TMTA63632 harboring blaIMP-68 (accession number AP019667) in a K. pneumoniae isolate obtained from a patient in Japan (Figs. 2 and 3) [23]. The plasmid incompatibility complex of pSL264 was IncL/M. Comparative analysis showed that the plasmids in E. cloacae SL 264 (pSL264), S. marcescens SL267 (pSL267) and K. aerogenes SL269 (pSL269) were completely identical. This plasmid, designated pSL264, was similar to other, previously identified plasmids, including pEB-1 in E. cloacae with 98.51% identity (KX230795) [24], pACM1 in Klebsiella oxytoca with 98.51% identity (KJ541681) [25], p51929_MCR_VIM in Citrobacter freundii with 98.51% identity (CP059429) [26], and pKp616_2 in K. pneumoniae with 98.52% identity (CP026497) [27]. However, the genetic environment surrounding blaIMP-1 in pSL264 was found to be unique.
Comparative analysis of plasmids highly similar to pSL264. pSL264 was 98.51% identical to the plasmid pEB-1 in E. cloacae (KX230795), 98.51% identical to pACM1 in Klebsiella oxytoca (KJ541681), 98.51% identical to p51929_MCR_VIM in Citrobacter freundii CP059429), and 98.52% identical to pKp616_2 in K. pneumoniae (CP026497)
Transferability of plasmid pSL264 harboring bla IMP-1
Conjugation experiments showed that pSL264 could transfer from S. marcescens to E. cloacae and K. aerogenes, from E. cloacae to K. aerogenes and from K. aerogenes to E. cloacae. In contrast, pSL264 could not transfer from E. cloacae or K. aerogenes to S. marcescens (Fig. 4 and Table 2).
Discussion
This study indicated that the plasmid, pSL264, transferred among three species of bacteria, E. cloacae, K. aerogenes and S. marcescens, in a single patient. However, our conjugation experiments indicated that pSL264 did not equally transfer among these species. Rather, it transferred from S. marcescens to E. cloacae and K. aerogenes, but not from E. cloacae or K. aerogenes to S. marcescen, suggesting that, in this patient, pSL264 was initially present in S. marcescens and subsequently spread to the other species. Although transferability rate in vitro was not that high, treatment with carbapenem for more than 28 days may have triggered the transfer of resistance.
This study had several limitations. First, this was a single center study. The transferability of pSL264 may not be general, and external validation may be required. Second, although in vitro conjugation experiments suggested that S. marcescens initially harbored pSL264, E. cloacae was isolated from this patient earlier than the other two species.
IncL/M plasmids harboring blaIMPs and blaNDMs in Enterobacteriaceae may be emerging and spreading, especially in Asian countries. An IncL/M plasmid was first reported in a multidrug-resistant strain of Morganella morganii isolated in South Africa in 1972 [28]. These plasmids are now commonly identified among environmental and clinical isolates [29, 30]. Because these plasmids are carriers of genes encoding carbapenemases, mostly OXA-48 carbapenemases, they can be regarded as a public health threat [23, 28, 30,31,32,33,34,35,36,37,38,39,40,41,42,43,44].
To our knowledge, this is the first report of an IncL/M plasmid harboring blaIMP-1 in three species of Enterobacteriaceae isolated from an individual patient. This plasmid may have spread clonally in the hospital environment, suggesting the need to assess the presence of Enterobacteriaceae harboring blaIMP-1 in medical settings in Japan.
Conclusion
This study found that pSL264 harboring blaIMP-1 could easily transfer among species of Enterobacteriaceae in a patient during long-term antimicrobial treatment. Molecular and genomic analyses of plasmids may reveal the horizontal transmission of plasmids in CPE.
Availability of data and materials
The chromosome and two plasmid sequences of E. cloacae SL264 have been deposited at GenBank under accession number AP024913, AP024914 and AP024915, respectively. The chromosome and the plasmid sequences of S. marcescens SL267 were AP024916 and AP024916, respectively. The chromosome and two plasmid sequences of K. aerogenes SL269 have been deposited at GenBank under Accession Number AP024918, AP024919 and AP024920, respectively.
References
Paterson DL, Doi Y. A step closer to extreme drug resistance (XDR) in gram-negative bacilli. Clin Infect Dis. 2007;45:1179–81.
Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerging Infect Dis. 2011;17:1791–8.
Bush K. New beta-lactamases in gram-negative bacteria: diversity and impact on the selection of antimicrobial therapy. Clin Infect Dis. 2001;32:1085–9.
Watanabe M, Iyobe S, Inoue M, Mitsuhashi S. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991;35:147–51.
Kitao T, Tada T, Tanaka M, Narahara K, Shimojima M, Shimada K, et al. Emergence of a novel multidrug-resistant Pseudomonas aeruginosa strain producing IMP-type metallo-β-lactamases and AAC(6’)-Iae in Japan. Int J Antimicrob Agents. 2012;39:518–21.
Tada T, Miyoshi-Akiyama T, Shimada K, Shiroma A, Nakano K, Teruya K, et al. A carbapenem-resistant Pseudomonas aeruginosa isolate harboring two copies of blaIMP-34 encoding a metallo-β-lactamase. PLoS ONE. 2016;11:e0149385.
Tada T, Nhung PH, Miyoshi-Akiyama T, Shimada K, Tsuchiya M, Phuong DM, et al. Multidrug-resistant sequence type 235 Pseudomonas aeruginosa clinical isolates producing IMP-26 with increased carbapenem-hydrolyzing activities in Vietnam. Antimicrob Agents Chemother. 2016;60:6853–8.
Carattoli A. Plasmids and the spread of resistance. Int J Med Microbiol. 2013;303:298–304.
Tada T, Uechi K, Nakasone I, Nakamatsu M, Satou K, Hirano T, et al. Emergence of IncX4 plasmids encoding mcr-1 in a clinical isolate of Klebsiella pneumoniae in Japan. Int J Infect Dis. 2018;75:98–100.
Wang Y, Tong MK, Chow KH, Cheng VCC, Tse CWS, Wu AKL, et al. Occurrence of highly conjugative IncX3 epidemic plasmid carrying blaNDM in Enterobacteriaceae isolates in geographically widespread areas. Front Microbiol. 2018;9:2272.
Paskova V, Medvecky M, Skalova A, Chudejova K, Bitar I, Jakubu V, et al. Characterization of NDM-encoding plasmids from enterobacteriaceae recovered from Czech hospitals. Front Microbiol. 2018;9:1549.
Matsumura Y, Peirano G, Bradford PA, Motyl MR, DeVinney R, Pitout JDD. Genomic characterization of IMP and VIM carbapenemase-encoding transferable plasmids of Enterobacteriaceae. J Antimicrob Chemother. 2018;73:3034–8.
Hardiman CA, Weingarten RA, Conlan S, Khil P, Dekker JP, Mathers AJ, et al. Horizontal transfer of carbapenemase-encoding plasmids and comparison with hospital epidemiology data. Antimicrob Agents Chemother. 2016;60:4910–9.
Göttig S, Gruber TM, Stecher B, Wichelhaus TA, Kempf VAJ. In vivo horizontal gene transfer of the carbapenemase OXA-48 during a nosocomial outbreak. Clin Infect Dis. 2015;60:1808–15.
Chudejova K, Kraftova L, Mattioni Marchetti V, Hrabak J, Papagiannitsis CC, Bitar I. Genetic plurality of OXA/NDM-encoding features characterized from Enterobacterales recovered from Czech hospitals. Front Microbiol. 2021;12:641415.
Papagiannitsis CC, Dolejska M, Izdebski R, Dobiasova H, Studentova V, Esteves FJ, et al. Characterization of pKP-M1144, a novel ColE1-like plasmid encoding IMP-8, GES-5, and BEL-1 β-lactamases, from a Klebsiella pneumoniae sequence type 252 isolate. Antimicrob Agents Chemother. 2015;59:5065–8.
Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. CLSI M100-S25. Clinical and Laboratory Standards Institute, Wayne, PA. 7th ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2015.
Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595.
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75.
Anjum MF, Duggett NA, AbuOun M, Randall L, Nunez-Garcia J, Ellis RJ, et al. Colistin resistance in Salmonella and Escherichia coli isolates from a pig farm in Great Britain. J Antimicrob Chemother. 2016;71(8):2306–13.
Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50:1355–61.
Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75:3491.
Kubota H, Suzuki Y, Okuno R, Uchitani Y, Ariyoshi T, Takemura N, et al. IMP-68, a novel IMP-type metallo-β-lactamase in imipenem-susceptible Klebsiella pneumoniae. mSphere. 2019. https://doi.org/10.1128/mSphere.00736-19.
Cuzon G, Bogaerts P, Bauraing C, Huang T-D, Bonnin RA, Glupczynski Y, et al. Spread of plasmids carrying multiple GES variants. Antimicrob Agents Chemother. 2016;60:5040–3.
Preston KE, Hitchcock SA, Aziz AY, Tine JA. The complete nucleotide sequence of the multi-drug resistance-encoding IncL/M plasmid pACM1. Plasmid. 2014;76:54–65.
Bitar I, Papagiannitsis CC, Kraftova L, Chudejova K, Mattioni Marchetti V, Hrabak J. Detection of five mcr-9-carrying Enterobacterales isolates in four Czech hospitals. mSphere. 2020;5:01008–20.
Wyres KL, Hawkey J, Hetland MAK, Fostervold A, Wick RR, Judd LM, et al. Emergence and rapid global dissemination of CTX-M-15-associated Klebsiella pneumoniae strain ST307. J Antimicrob Chemother. 2019;74:577–81.
Hedges RW, Datta N, Coetzee JN, Dennison S. R factors from Proteus morganii. J Gen Microbiol. 1973;77:249–59.
Poirel L, Bonnin RA, Nordmann P. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob Agents Chemother. 2012;56:559–62.
Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother. 2009;53:2227–38.
Al-Baloushi AE, Pál T, Ghazawi A, Sonnevend A. Genetic support of carbapenemases in double carbapenemase producer Klebsiella pneumoniae isolated in the Arabian Peninsula. Acta Microbiol Immunol Hung. 2018;65:135–50.
Power K, Wang J, Karczmarczyk M, Crowley B, Cotter M, Haughton P, et al. Molecular analysis of OXA-48-carrying conjugative IncL/M-like plasmids in clinical isolates of Klebsiella pneumoniae in Ireland. Microb Drug Resist. 2014;20:270–4.
Partridge SR, Ginn AN, Paulsen IT, Iredell JR. pEl1573 Carrying blaIMP-4, from Sydney, Australia, is closely related to other IncL/M plasmids. Antimicrob Agents Chemother. 2012;56:6029–32.
Di Pilato V, Arena F, Giani T, Conte V, Cresti S, Rossolini GM. Characterization of pFOX-7a, a conjugative IncL/M plasmid encoding the FOX-7 AmpC-type β-lactamase, involved in a large outbreak in a neonatal intensive care unit. J Antimicrob Chemother. 2014;69:2620–4.
Wang Y, Lo W-U, Lai EL, Chow K-H, Ho P-L. Complete sequence of the multidrug-resistant IncL/M plasmid pIMP-HB623 Cocarrying bla IMP-34 and fosC2 in an Enterobacter cloacae strain associated with medical travel to China. Antimicrob Agents Chemother. 2015;59:5854–6.
Espedido BA, Steen JA, Ziochos H, Grimmond SM, Cooper MA, Gosbell IB, et al. Whole genome sequence analysis of the first Australian OXA-48-producing outbreak-associated Klebsiella pneumoniae isolates: the resistome and in vivo evolution. PLoS ONE. 2013;8:e59920.
Shigemoto N, Kayama S, Kuwahara R, Hisatsune J, Kato F, Nishio H, et al. A novel metallo-β-lactamase, IMP-34, in Klebsiella isolates with decreased resistance to imipenem. Diagn Microbiol Infect Dis. 2013;76:119–21.
Xie L, Dou Y, Zhou K, Chen Y, Han L, Guo X, et al. Coexistence of blaOXA-48 and truncated blaNDM-1 on different plasmids in a Klebsiella pneumoniae Isolate in China. Front Microbiol. 2017;8:133.
Berger S, Alauzet C, Aissa N, Hénard S, Rabaud C, Bonnet R, et al. Characterization of a new blaOXA-48-carrying plasmid in Enterobacteriaceae. Antimicrob Agents Chemother. 2013;57:4064–7.
Rimoldi SG, Gentile B, Pagani C, Di Gregorio A, Anselmo A, Palozzi AM, et al. Whole genome sequencing for the molecular characterization of carbapenem-resistant Klebsiella pneumoniae strains isolated at the Italian ASST Fatebenefratelli Sacco Hospital, 2012–2014. BMC Infect Dis. 2017;17:666.
Ho PL, Lo WU, Yeung MK, Lin CH, Chow KH, Ang I, et al. Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS ONE. 2011;6:e17989.
Bonnin RA, Nordmann P, Carattoli A, Poirel L. Comparative genomics of IncL/M-Type plasmids: evolution by acquisition of resistance genes and insertion sequences. Antimicrob Agents Chemother. 2013;57:674–6.
Bryant KA, Van Schooneveld TC, Thapa I, Bastola D, Williams LO, Safranek TJ, et al. KPC-4 Is encoded within a truncated Tn4401 in an IncL/M plasmid, pNE1280, isolated from Enterobacter cloacae and Serratia marcescens. Antimicrob Agents Chemother. 2013;57:37–41.
Guo L, An J, Ma Y, Ye L, Luo Y, Tao C, et al. Nosocomial outbreak of OXA-48-producing Klebsiella pneumoniae in a Chinese Hospital: clonal transmission of ST147 and ST383. PLoS ONE. 2016;11:e0160754.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the Research Program on Emerging and Re-emerging Infectious Diseases from Japan Agency for Medical Research and Development (Grant Number: 21fk0108604h0701). The funding organization had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
The manuscript was seen and approved by all the authors and is not under consideration elsewhere. All the authors contributed to the work in this study. NM collected clinical data and wrote the initial draft of the manuscript; TT designed this study; SO helped perform the conjugation experiments; KK performed the drug susceptibility profiles and data analysis; TK supervised this study and edited the manuscript; and YU helped perform genomic analyses and supervised this study. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participant
This study was approved by the Institutional Review Board of St. Luke’s International University in Tokyo, Japan (Number: 20-R151) and by the Biosafety Committee, Juntendo University (Approval Number BSL2/29-1). Individual informed consent was waived by the ethics committee listed above because this study used currently existing sample collected during the course of routine medical care and did not pose any additional risks to the patients. Informed consent about study participation was officially announced by mail and poster. All patient data were anonymized prior to the analysis.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Mori, N., Tada, T., Oshiro, S. et al. A transferrable IncL/M plasmid harboring a gene encoding IMP-1 metallo-β-lactamase in clinical isolates of Enterobacteriaceae. BMC Infect Dis 21, 1061 (2021). https://doi.org/10.1186/s12879-021-06758-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-021-06758-5