Abstract
Background
Several studies suggested pancreatic stone protein (PSP) as a promising biomarker to predict mortality among patients with severe infection. The objective of the study was to evaluate the performance of PSP in predicting intensive care unit (ICU) mortality and infection severity among critically ill adults admitted to the hospital for infection.
Methods
A systematic search across Cochrane Central Register of Controlled Trials and MEDLINE databases (1966 to February 2022) for studies on PSP published in English using ‘pancreatic stone protein’, ‘PSP’, ‘regenerative protein’, ‘lithostatin’ combined with ‘infection’ and ‘sepsis’ found 46 records. The search was restricted to the five trials that measured PSP using the enzyme-linked immunosorbent assay technique (ELISA). We used Bayesian hierarchical regression models for pooled estimates and to predict mortality or disease severity using PSP, C-Reactive Protein (CRP) and procalcitonin (PCT) as main predictor. We used statistical discriminative measures, such as the area under the receiver operating characteristic curve (AUC) and classification plots.
Results
Among the 678 patients included, the pooled ICU mortality was 17.8% (95% prediction interval 4.1% to 54.6%) with a between-study heterogeneity (I-squared 87%). PSP was strongly associated with ICU mortality (OR = 2.7, 95% credible interval (CrI) [1.3–6.0] per one standard deviation increase; age, gender and sepsis severity adjusted OR = 1.5, 95% CrI [0.98–2.8]). The AUC was 0.69 for PSP 95% confidence interval (CI) [0.64–0.74], 0.61 [0.56–0.66] for PCT and 0.52 [0.47–0.57] for CRP. The sensitivity was 0.96, 0.52, 0.30 for risk thresholds 0.1, 0.2 and 0.3; respective false positive rate values were 0.84, 0.25, 0.10.
Conclusions
We found that PSP showed a very good discriminative ability for both investigated study endpoints ICU mortality and infection severity; better in comparison to CRP, similar to PCT. Combinations of biomarkers did not improve their predictive ability.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
The early recognition of patients with severe infections and potentially unfavorable outcome is critical to improve mortality in sepsis, as patients at high-risk of death might benefit from individualized care and advanced support [1]. Biomarkers are increasingly being used to target personalized care and precision medicine in various clinical settings [2,3,4,5], including for the management of sepsis [6, 7]. C-reactive protein (CRP) and procalcitonin (PCT) are broadly used to stratify infection according to disease severity and potential outcome despite their poor performance for that purpose [8,9,10,11,12,13]. Other biomarkers have been proposed, but their place in clinical practice is not established [14,15,16].
Pancreatic stone protein (PSP) has recently emerged as a promising biomarker of infection [17].
PSP is a globular polypeptide adopting a fold described for C-type lectins with a diverse range of functions, including signalling receptors in homeostasis and innate immunity, playing a crucial role in inflammatory response and leukocyte and platelet trafficking. It is mostly synthesized by the pancreas and the intestine with increasing blood levels early in the context of sepsis [17]. The point-of-care (POC) machines for bedside analysis only need a drop of whole blood to deliver results within few minutes [17].
Over the last two decades, PSP has been thoroughly evaluated in various medico-surgical patient populations and multiple clinical settings, especially in emergency rooms (ER), burn and intensive care units (ICUs) [18,19,20,21,22,23]. Several studies, including a recent meta-analysis [24], conducted in adults, children and neonates investigated the capacity of PSP to diagnose infection [20,21,22], characterize disease severity [19, 23] and predict outcome of patients with sepsis [19, 23, 25,26,27].
Here, we perform an individual patient level meta-analysis to evaluate the ability of PSP to predict patients with poor outcome and/or severe disease and report classification plots with continuous risk thresholds to support clinical decision-making based on current recommendations for predictions models [34].
Methods
Search strategy and selection criteria
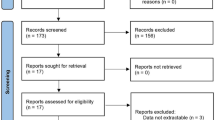
A systematic literature search was performed across the Cochrane Central Register of Controlled Trials (CENTRAL and MEDLINE (1966 to February 2022) databases using “pancreatic stone protein”, “PSP”, “regenerative protein”, “infection”, “sepsis”, “lithostatin” as keywords and/or MeSH Terms. The search strategy was prepared according to PRISMA individual patient data guidelines (Supplemental Tables 1 and 2) [28]. The search was restricted to original human clinical trials on PSP/reg published in English before February 2022 that evaluated the performance of PSP for the assessment of the severity of infection as well as for predicting its outcome among unselected adult patients upon their admission to the ED or the ICU. The search was further restricted to studies that determined PSP levels in blood using the enzyme-linked immunosorbent assay technique (ELISA) developed and described by Rolf Graf et al. [20, 29], to impede calculation limitations when plotting equal PSP levels when using different analysing methods. Paediatric trials and autopsy studies were excluded. The definitions of infection used in each of the eligible studies are presented in Supplemental Table 3.
Two reviewers (JP and YAQ) independently assessed trial eligibility based on titles, abstracts, full-text reports, and further information from investigators as needed (Fig. 1). Study protocols and unedited databases containing anonymized individual patient data were obtained from investigators of all eligible trials.
The study was registered on Prospero (#CRD42022308207). The Cantonal Ethical Committee of the State of Bern (#2018-01356_V2.1_25.2.2022) reviewed and approved the meta-analysis research protocol while the respective ethical committees already approved all individual studies.
Assessment of data validity
All raw data were received from their principal investigators with patient specific anonymized ID and contained at least the following information: age, gender, Sequential Organ Failure Assessment (SOFA) Score and blood levels of PSP, CRP and PCT upon admission, days to death and ICU mortality. Data from each eligible study were first checked for duplicates and second against reported results. Queries were resolved with the principal investigator, trial data manager, or statistician whenever indicated.
Study objectives
The primary objective of the study was to evaluate the diagnostic accuracy of PSP in predicting ICU mortality and compare it to CRP and PCT. The secondary objectives were: (i) to evaluate PSP ability to predict disease severity and compare it to CRP and PCT, and (ii) to explore whether different combinations of the three biomarkers further improve the prediction of ICU-mortality and disease severity.
Study outcomes
Our primary endpoint was ICU mortality. Secondary outcomes were based on disease severity risk stratification on SOFA score upon admission: (i) non-complicated infection (patients with SOFA score ≤ 1; (ii) sepsis (patients with SOFA score ≥ 2) and (iii) septic shock (patients with SOFA score ≥ 2 and need for vasoactive drugs). We used the combined endpoint (sepsis and septic shock) as secondary outcome.
Confounders
We adjusted all outcomes for age and sex. For the primary outcome ICU-mortality, we additionally adjusted for sepsis severity (mild moderate infection/infection, sepsis, septic shock).
Statistical analysis
We described the study population by counts and percentages, median and interquartile range. Missing PSP, CRP or PCT measurements were replaced by median values within each study, because of the low missing value proportion: Percentage of missing values per study ranged from 0.4% for CRP/PCT to 5.6% for PSP (Supplemental Table 4). For adjusted analyses, three missing age values were replaced by the median value of the corresponding study.
We followed the meta-analytic approach used by Prazak et al. [24] and described in Steyerberg et al. [30]. Briefly, we evaluated three different models: (i) a random effect-random slope (RERS) model (random intercept on study and biomarkers as random slopes including a fixed effect on biomarkers for population mean interpretation of the random intercept and slope [31]); (ii) a random effect (RE) model (random intercept on study and fixed biomarker effect); and (iii) a fixed effect (FE) model (fixed biomarker effects without any patient clustering information). We compared models using the Akaike information criterion (AIC) and log-likelihoods. Because of the small number of studies and convergence issues of frequentist random effects models, we used Bayesian hierarchical logistic regression models. We used centered Gaussian priors with a standard deviation of 2.5 for intercept and biomarker effects [32]. For the centered multivariate Gaussian distributed random effects we used a Lewandowski-Kurowicka-Joe prior with a regularization parameter set to 1, a concentration parameter set to 1 and a unit-exponential prior on the scale parameters for the decomposition of the correlation matrix [2]. We used unadjusted models (using only biomarker values as predictors) and adjusted models (biomarker values and all confounding variables) reporting odds ratio with 95% credible intervals (CrI). Biomarker measurements were standardized (centered and divided by population standard deviation) and age centered and expressed as a 10-year increase. We reported study-specific outcome estimates and 95% CrI as well as between-study standard deviation and I-squared. 95% prediction intervals (PI) were calculated from the overall intercept plus a centered Gaussian distributed random variable with a standard deviation equal to the estimated between-study standard deviation. We reported AUC values with 95% confidence intervals (CIs), positive and negative predictive values, and classification plots [33]. A specific risk threshold cutoff was computed based on Youden’s index [34]. All analyses were performed in R version 4.1.2 [27]. Bayesian analyses were implemented in the Stan R interface [2] using 4 Markov chains with 1,000 warmup iterations per chain and 2,000 total iterations per chain.
Results
Study selection
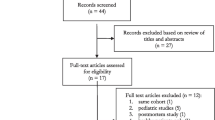
Among the 48 records published before February 2022 and identified through the literature search, 46 full texts were further assessed for eligibility. 24 records were excluded based on review of title and abstracts. Mainly due to lack of measured biomarkers on admission or addressing a pediatric patient population, only five of the remaining 22 observational studies were included into the final analysis (Fig. 1 ; Table 1). Individual patient data from all patients were used for the evaluation of the primary endpoint ‘ICU mortality’ (Table 2). For the assessment of the secondary endpoint predicting disease severity, the studies of Que et al. [23] and Guadiana-Romualdo et al. (2019) [35] were excluded, since those studies only included patients with severe sepsis or septic shock (Fig. 1; Table 1).
Analysis population
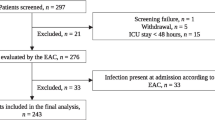
We considered 678 patients in the study; 64% were male with a median age of 65 (Table 2). The biomarkers were measured on 549 patients admitted to ICUs and on 129 admitted to the emergency room. The distributions of the three biomarkers by study disease severity are shown in supplement (Supplemental Figs 1 and 2).
ICU mortality
The observed crude overall ICU mortality was 22% (149 out of 678 included patients). Model performance was best for a RERS models based on AIC (Supplemental Table 5). The pooled overall estimate from a RERS model was 17.8%, 95% CrI (9.1–31.5%) with a 95% PI ranging from 4.1–54.6% with a substantial heterogeneity between studies (I-squared 87%), (Fig. 2).
PSP was strongly associated with ICU mortality (OR = 2.7, 95% CrI [1.3–6.0] per one SD increase), even after adjustment for age, gender and sepsis severity (OR = 1.5, 95% CrI [0.98–2.83], Supplemental Fig. 3). The AUC from an unadjusted RERS model was 0.69 [95%CI 0.64–0.74]. We identified a PSP cut-off value of 133.6 ng/ml based on Youden index at a risk threshold at 13% with positive (PPV, 0.32, 95%CI [0.27–0.36]) and negative (NPV, 0.90, 95%CI [0.87–0.93]) predictive values using PSP (Table 3). Calibration plots showed that the sensitivity for PSP was 0.96, 0.52, 0.30 for risk thresholds 10%, 20% and 30%; respective false positive rate values were 0.84, 0.25, 0.10 (Fig. 3). Similar analyses were performed for CRP and PCT. Combining biomarkers in all different models evaluated did not increase the discriminative performance of PSP (Supplemental Fig. 4; Supplemental Table 6).
Infection severity
PSP was higher in patients with sepsis/septic shock compared to those with mild infections and strongly associated with the combined endpoint of sepsis/septic shock in both unadjusted (OR = 11.4, 95% CrI [2.1–54.5]; per one SD increase and age–gender adjusted models (OR = 11.4, 95% CrI [1.9–48.9]), (Supplemental Fig. 5). For the secondary combined outcome of sepsis and septic shock we estimated a pooled overall percentage of 79.9%, with a 95% PI ranging from 5.5% to 99.6% with a considerable heterogeneity between studies (I-squared 93%), (Fig. 4).
Risk thresholds based on Youden index to discriminate mild infection form severe infection/septic shock were 61.7 ng/ml for PSP, 125.9 mg/l for CRP and 1.1 ng/ml for PCT (Fig. 5). Using those, PSP (AUC 0.80, 95%CI [0.75–0.85]) and PCT (AUC 0.79, 95%CI [0.74–0.84]) performed better that CRP (in stratifying patient according to infection severity: AUC was lowest for CRP (AUC 0.56, 95%CI [0.50–0.63]). PPV was the highest for PCT (0.87, 95%CI [0.81–0.92] and NPV for PSP (0.67, 95%CI [0.58–0.75]) (Supplemental Table 6). Discriminative performance (as measured by AUC) did not improve when biomarkers where combined (Supplemental Fig. 6; Supplemental Table 7).
Discussion
We analyzed individual patient level data from five studies that measured PSP using the enzyme-linked immunosorbent assay technique investigating the diagnostic accuracy of PSP on ICU mortality and infection severity. Our results suggest that PSP has a very good discriminative ability, higher than CRP and comparable to PCT. To the best of our knowledge, the present study is the first meta-analysis of its kind using actual datasets from different studies on this very topic.
Correctly identifying patients suffering from severe sepsis or septic shock and predicting ICU mortality is key when treating patients with infection not only to rapidly stabilize the patient’s condition and positively influence outcome, but also to allocate an adequate amount of resources. It is also important for identification of appropriate patients for enrollment in trials of sepsis interventions. Current clinical scoring systems lack sensitivity and specificity to guide decisions and prognostication upon admission [36,37,38]. Despite their large use for comparing severity and predicting mortality across ICU patient populations, common ICU severity scores such as Acute Physiology and Chronic Health Evaluation (APACHE II) and Simplified Acute Physiology Score (SAPS II) are not designed to recognize and discriminate between individual outcomes [39]. Recently, the National Early Warning Score (NEWS) [40] has emerged as valuable tool to predict sepsis-related outcomes upon admission [41] or after ICU transfer [42]. Nowadays, NEWS has been incorporated almost universally in the UK in the patient management [43].
Besides their application to diagnose infection and assess the response to therapy, biomarkers are also increasingly being used to stratify patients according to their risk profiles and to predict sepsis-related outcomes [44]. For instance, certain blood transcriptomics of gene panels might accurately predict patient outcome after burn [45] or blunt trauma [46] and identify those at risk of developing infection in the course of recovery. On a larger scale, the performance of the widely available classical biomarkers CRP, PCT as predictors of adverse outcomes still remain controversial [47, 48].
The present study is the first individual patient level meta-analysis that systematically evaluates the performance of PSP in predicting infection severity and outcome in patients upon admission to ICU or ER. PSP demonstrated better predictive ability for ICU mortality in comparison to canonical biomarkers of infection as CRP, but similar to PCT. In addition, PSP could reliably stratify patients according to infection severity. Altogether, our data suggest that PSP could be used as a prognostic biomarker in such patients and support precision medicine in the management of infections and sepsis [49].
Better information on patients’ individual risk profile and outcome upon the admission to the ER or the ICU should assist healthcare givers and clinicians in their triage decision to make timely allocation of resources and therapeutic options. Correct identification of high-urgency patients avoids delays in the initiation of sepsis management, while reliable classification of low-urgency patients improve efficiency in the ER patient flow. Such approaches have been successfully evaluated in specific clinical settings such as urinary tract infections [50] as well as in the unselected patient populations (within the TRIAGE study) [51]. One advantage of PSP over other blood biomarker is the availability of a POC diagnostic tests using nanofluid technology, enabling rapid quantification of PSP at the bedside [17, 26, 52].
Our study has several strengths. First, we received individual patient level data from the eligible studies, which allowed us to model our study endpoints and biomarkers on patient level. Second, the original studies were performed in different centers across Europe and covered two clinical settings, including ER and ICU, which make the results more generalizable. Finally, the use of classification plots in contrast to conventional ROC allows for a direct visualization of the model’s discriminative ability enabling the clinicians to choose the threshold value according to the therapeutic question. A single threshold based on Youden index might be suboptimal from a clinical view, such that classification plots are a helpful tool to support clinicians in decision making. The main limitations of the meta-analysis are the relatively small numbers of included studies and the exclusion of newer ones performed using the recently available POC technology. Direct comparison with the previous ELISA technique with which all PSP levels were measured, is possible as POC PSP levels approximately equals 4.6 × previous ELISA ng/ml + 30 ng/ml [52].
Conclusions
In conclusion, the present study confirms that PSP is a promising biomarker to predict sepsis-related outcome and estimate infection severity upon hospital and/or ICU admission. However, further prospective studies are needed to confirm its utility and safety in the daily clinical use.
Availability of data and material
The Corresponding author has acces to all data included into the analysis. Requests should be submitted to the corresponding author in the first instance.
Abbreviations
- CI:
-
Confidence interval
- CrI:
-
Credible interval
- ER:
-
Emergency room
- CRP:
-
C-reactive protein
- ICU:
-
Intensive care unit
- PCT:
-
Procalcitonin
- PSP:
-
Pancreatic stone protein
References
Rhodes A, Evans LE, Alhazzani W, Levy MM, Antonelli M, Ferrer R, Kumar A, Sevransky JE, Sprung CL, Nunnally ME, et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock: 2016. Intensive Care Med. 2017;43:304–77.
Greer O, Shah NM, Sriskandan S, Johnson MR. Sepsis: precision-based medicine for pregnancy and the puerperium. Int J Mol Sci. 2019;20:5388.
Aletaha D. Precision medicine and management of rheumatoid arthritis. J Autoimmun. 2020;110: 102405.
Ho D, Quake SR, McCabe ERB, Chng WJ, Chow EK, Ding X, Gelb BD, Ginsburg GS, Hassenstab J, Ho CM, et al. Enabling technologies for personalized and precision medicine. Trends Biotechnol. 2020;38:497–518.
Seppälä TT, Zimmerman JW, Suri R, Zlomke H, Ivey GD, Szabolcs A, Shubert CR, Cameron JL, Burns WR, Lafaro KJ, et al. Precision medicine in pancreatic cancer: patient-derived organoid pharmacotyping is a predictive biomarker of clinical treatment response. Clin Cancer Res. 2022;28:3296–307.
van Engelen TSR, Wiersinga WJ, Scicluna BP, van der Poll T. Biomarkers in sepsis. Crit Care Clin. 2018;34:139–52.
Singer M. Sepsis: personalization v protocolization? Crit care (London, England). 2019;23:127.
Albrich WC, Harbarth S. Pros and cons of using biomarkers versus clinical decisions in start and stop decisions for antibiotics in the critical care setting. Intens Care Med. 2015;41:1739–51.
Schuetz P, Briel M, Christ-Crain M, Stolz D, Bouadma L, Wolff M, Luyt CE, Chastre J, Tubach F, Kristoffersen KB, et al. Procalcitonin to guide initiation and duration of antibiotic treatment in acute respiratory infections: an individual patient data meta-analysis. Clin infect Dis. 2012;55:651–62.
Schuetz P, Chiappa V, Briel M, Greenwald JL. Procalcitonin algorithms for antibiotic therapy decisions: a systematic review of randomized controlled trials and recommendations for clinical algorithms. Arch Intern Med. 2011;171:1322–31.
Simon L, Gauvin F, Amre DK, Saint-Louis P, Lacroix J. Serum procalcitonin and C-reactive protein levels as markers of bacterial infection: a systematic review and meta-analysis. Clin Infect Dis. 2004;39:206–17.
Tang BM, Eslick GD, Craig JC, McLean AS. Accuracy of procalcitonin for sepsis diagnosis in critically ill patients: systematic review and meta-analysis. Lancet Infect Dis. 2007;7:210–7.
Wacker C, Prkno A, Brunkhorst FM, Schlattmann P. Procalcitonin as a diagnostic marker for sepsis: a systematic review and meta-analysis. Lancet Infect Dis. 2013;13:426–35.
Pierrakos C, Velissaris D, Bisdorff M, Marshall JC, Vincent JL. Biomarkers of sepsis: time for a reappraisal. Crit Care (London, England). 2020;24:287.
Pierrakos C, Vincent JL. Sepsis biomarkers: a review. Crit Care (London, England). 2010;14:R15.
Rhee C, Kadri SS, Danner RL, Suffredini AF, Massaro AF, Kitch BT, Lee G, Klompas M. Diagnosing sepsis is subjective and highly variable: a survey of intensivists using case vignettes. Crit Care (London, England). 2016;20:89.
Eggimann P, Que YA, Rebeaud F. Measurement of pancreatic stone protein in the identification and management of sepsis. Biomark Med. 2019;13:135–45.
Garcia de Guadiana-Romualdo L, Berger M, Jimenez-Santos E, Rebollo-Acebes S, Jimenez-Sanchez R, Esteban-Torrella P, Hernando-Holgado A, Ortin-Freire A, Albaladejo-Oton MD. Pancreatic stone protein and soluble CD25 for infection and sepsis in an emergency department. Eur J Clin Investig 2017, 47:297–304.
Gukasjan R, Raptis DA, Schulz HU, Halangk W, Graf R. Pancreatic stone protein predicts outcome in patients with peritonitis in the ICU. Crit Care Med. 2013;41:1027–36.
Keel M, Harter L, Reding T, Sun LK, Hersberger M, Seifert B, Bimmler D, Graf R. Pancreatic stone protein is highly increased during posttraumatic sepsis and activates neutrophil granulocytes. Crit Care Med. 2009;37:1642–8.
Klein HJ, Csordas A, Falk V, Slankamenac K, Rudiger A, Schonrath F. Rodriguez Cetina Biefer H, Starck CT, Graf R: Pancreatic stone protein predicts postoperative infection in cardiac surgery patients irrespective of cardiopulmonary bypass or surgical technique. PLoS One. 2015;10: e0120276.
Llewelyn MJ, Berger M, Gregory M, Ramaiah R, Taylor AL, Curdt I, Lajaunias F, Graf R, Blincko SJ, Drage S, et al. Sepsis biomarkers in unselected patients on admission to intensive or high-dependency care. Crit Care (London, England). 2013;17:R60.
Que YA, Guessous I, Dupuis-Lozeron E, de Oliveira CRA, Oliveira CF, Graf R, Seematter G, Revelly JP, Pagani JL, Liaudet L, et al. Prognostication of mortality in critically Ill patients with severe infections. Chest. 2015;148:674–82.
Prazak J, Irincheeva I, Llewelyn MJ, Stolz D, García de Guadiana Romualdo L, Graf R, Reding T, Klein HJ, Eggimann P, Que YA. Accuracy of pancreatic stone protein for the diagnosis of infection in hospitalized adults: a systematic review and individual patient level meta-analysis. Crit Care (London, England) 2021, 25:182.
Boeck L, Graf R, Eggimann P, Pargger H, Raptis DA, Smyrnios N, Thakkar N, Siegemund M, Rakic J, Tamm M, et al. Pancreatic stone protein: a marker of organ failure and outcome in ventilator-associated pneumonia. Chest. 2011;140:925–32.
Pugin J, Daix T, Pagani JL, Morri D, Giacomucci A, Dequin PF, Guitton C, Que YA, Zani G, Brealey D, et al. Serial measurement of pancreatic stone protein for the early detection of sepsis in intensive care unit patients: a prospective multicentric study. Crit Care (London, England). 2021;25:151.
Rodríguez Rojas C, García de Guadiana-Romualdo L, Morán Sánchez S, Prazak J, Algara Soriano V, Que YA, Benninga R, Albaladejo-Otón MD: Role of pancreatic stone protein as an early biomarker for risk stratification of acute pancreatitis. Dig Dis Sci 2022, 67:3275-83.
Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6: e1000097.
Yang J, Li L, Raptis D, Li X, Li F, Chen B, He J, Graf R, Sun Z. Pancreatic stone protein/regenerating protein (PSP/reg): a novel secreted protein up-regulated in type 2 diabetes mellitus. Endocrine. 2015;48:856–62.
Steyerberg EW, Nieboer D, Debray TPA, van Houwelingen HC. Assessment of heterogeneity in an individual participant data meta-analysis of prediction models: an overview and illustration. Stat Med. 2019;38:4290–309.
Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67:1–48.
Gelman A, Hill J, Vehtari A. Regression and Other Stories. Cambridge: Cambridge University Press; 2020.
Verbakel JY, Steyerberg EW, Uno H, De Cock B, Wynants L, Collins GS, Van Calster B. ROC curves for clinical prediction models part 1. ROC plots showed no added value above the AUC when evaluating the performance of clinical prediction models. J Clin Epidemiol. 2020;126:207–16.
Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3:32–5.
García de Guadiana-Romualdo L, Albaladejo-Otón MD, Berger M, Jiménez-Santos E, Jiménez-Sánchez R, Esteban-Torrella P, Rebollo-Acebes S, Hernando-Holgado A, Ortín-Freire A, Trujillo-Santos J: Prognostic performance of pancreatic stone protein in critically ill patients with sepsis. Biomark Med 2019, 13:1469–80.
Churpek MM, Snyder A, Han X, Sokol S, Pettit N, Howell MD, Edelson DP. Quick sepsis-related organ failure assessment, systemic inflammatory response syndrome, and early warning scores for detecting clinical deterioration in infected patients outside the intensive care unit. Am J Respir Crit Care Med. 2017;195:906–11.
Freund Y, Lemachatti N, Krastinova E, Van Laer M, Claessens YE, Avondo A, Occelli C, Feral-Pierssens AL, Truchot J, Ortega M, et al. Prognostic accuracy of sepsis-3 criteria for in-hospital mortality among patients with suspected infection presenting to the emergency department. JAMA. 2017;317:301–8.
Raith EP, Udy AA, Bailey M, McGloughlin S, MacIsaac C, Bellomo R, Pilcher DV. Australian, New Zealand intensive care society centre for O, Resource E: prognostic accuracy of the SOFA score, SIRS criteria, and qSOFA score for in-hospital mortality among adults with suspected infection admitted to the intensive care unit. JAMA. 2017;317:290–300.
Vincent JL, Opal SM, Marshall JC. Ten reasons why we should NOT use severity scores as entry criteria for clinical trials or in our treatment decisions. Crit Care Med. 2010;38:283–7.
Smith GB, Prytherch DR, Meredith P, Schmidt PE, Featherstone PI. The ability of the National Early Warning Score (NEWS) to discriminate patients at risk of early cardiac arrest, unanticipated intensive care unit admission, and death. Resuscitation. 2013;84:465–70.
Redfern OC, Smith GB, Prytherch DR, Meredith P, Inada-Kim M, Schmidt PE. A comparison of the quick sequential (Sepsis-Related) organ failure assessment score and the national early warning score in non-ICU patients with/without infection. Crit Care Med. 2018;46:1923–33.
Uppanisakorn S, Bhurayanontachai R, Boonyarat J, Kaewpradit J. National early warning score (NEWS) at ICU discharge can predict early clinical deterioration after ICU transfer. J Crit Care. 2018;43:225–9.
Williams B. The national early warning score: from concept to NHS implementation. Clin Med (Lond). 2022;22:499–505.
Póvoa P, Coelho L, Dal-Pizzol F, Ferrer R, Huttner A, Conway Morris A, Nobre V, Ramirez P, Rouze A, Salluh J, et al. How to use biomarkers of infection or sepsis at the bedside: guide to clinicians. Intens Care Med. 2023;49:142–53.
Yan S, Tsurumi A, Que YA, Ryan CM, Bandyopadhaya A, Morgan AA, Flaherty PJ, Tompkins RG, Rahme LG. Prediction of multiple infections after severe burn trauma: a prospective cohort study. Ann Surg. 2015;261:781–92.
Tsurumi A, Flaherty PJ, Que Y-A, Ryan CM, Mendoza AE, Almpani M, Bandyopadhaya A, Ogura A, Dhole YV, Goodfield LF et al: Multi-biomarker prediction models for multiple infection episodes following blunt trauma. iScience 2020, 23:101659.
Arora S, Singh P, Singh PM, Trikha A. Procalcitonin levels in survivors and nonsurvivors of sepsis: systematic review and meta-analysis. Shock. 2015;43:212–21.
Velissaris D, Zareifopoulos N, Lagadinou M, Platanaki C, Tsiotsios K, Stavridis EL, Kasartzian DI, Pierrakos C, Karamouzos V. Procalcitonin and sepsis in the emergency department: an update. Eur Rev Med Pharmacol Sci. 2021;25:466–79.
Watkins RR, Bonomo RA, Rello J. Managing sepsis in the era of precision medicine: challenges and opportunities. Expert Rev Anti Infect Ther. 2022;20:871–80.
Litke A, Bossart R, Regez K, Schild U, Guglielmetti M, Conca A, Schäfer P, Reutlinger B, Mueller B, Albrich WC. The potential impact of biomarker-guided triage decisions for patients with urinary tract infections. Infection. 2013;41:799–809.
Schuetz P, Hausfater P, Amin D, Amin A, Haubitz S, Faessler L, Kutz A, Conca A, Reutlinger B, Canavaggio P, et al. Biomarkers from distinct biological pathways improve early risk stratification in medical emergency patients: the multinational, prospective, observational TRIAGE study. Crit Care (London, England). 2015;19:377.
Benninga R: Nanofluidic technology enables rapid quantification of pancreatic stone protein as an early biomarker of sepsis: method comparison of the abioSCOPE in-vitro diagnostic device. J Appl Labor Med p 28th AACC International CPOCT Symposium.
Funding
Open access funding provided by University of Bern. This analysis was supported by unrestricted institutional research funds to JP and YAQ.
Author information
Authors and Affiliations
Contributions
JP, YAQ, PE and AM designed the study. AM performed the statistical analysis. JP, PE, AM, PZ and YAQ wrote the manuscript. MJL, LGGR, RG, TG, collected the raw data and critically revised the design of the study and the manuscript. All authors reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
All authors have completed the ICMJE uniform disclosure form at https://www.icmje.org/coi_disclosure.pdf and declare: no support from any organisation for the submitted work. RG has received research grants from Gebert Rüf Foundation, in addition, Dr. Graf has a patent method for assaying sepsis and outcome in humans by detection of PSP/reg licensed to LASCCO. DS reports grants from Astra-Zeneca AG, Curetis AG, Boston Scientific, other from Astra-Zeneca AG, Novartis AG, GSK AG, Roche AG, Zambon, Pfizer and Schwabe Pharma AG, Vifor AG, outside the submitted work. PE has received research grants from Abionic outside the submitted work.
Ethics approval and consent to participate
The study was registered on Prospero (#CRD42022308207). The Cantonal Ethical Committee of the State of Bern (#2018-01356_V2.1_25.2.2022) reviewed and approved the meta-analysis research protocol while the respective ethical committees already approved all individual studies.
Consent for publication
Not applicable.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zuercher, P., Moser, A., Garcia de Guadiana-Romualdo, L. et al. Discriminative performance of pancreatic stone protein in predicting ICU mortality and infection severity in adult patients with infection: a systematic review and individual patient level meta-analysis. Infection 51, 1797–1807 (2023). https://doi.org/10.1007/s15010-023-02093-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s15010-023-02093-w