Abstract
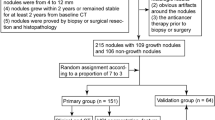
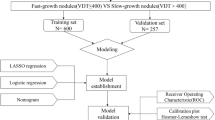
The objective is to evaluate the performance of computational image classification for indeterminate pulmonary nodules (IPN) chronologically detected by CT scan. Total 483 patients with 670 abnormal pulmonary nodules, who were taken chest thin-section CT (TSCT) images at least twice and resected as suspicious nodules in our hospital, were enrolled in this study. Nodular regions from the initial and the latest TSCT images were cut manually for each case, and approached by Python development environment, using the open-source cv2 library, to measure the nodular change rate (NCR). These NCRs were statistically compared with clinico-pathological factors, and then, this discriminator was evaluated for clinical performance. NCR showed significant differences among the nodular consistencies. In terms of histological subtypes, NCR of invasive adenocarcinoma (ADC) were significantly distinguishable from other lesions, but not from minimally invasive ADC. Only for cancers, NCR was significantly associated with loco-regional invasivity, p53-immunoreactivity, and Ki67-immunoreactivity. Regarding Epidermal Growth Factor Receptor gene mutation of ADC-related nodules, NCR showed a significant negative correlation. On staging of lung cancer cases, NCR was significantly increased with progression from pTis-stage 0 up to pT1b-stage IA2. For clinical shared decision-making (SDM) whether urgent resection or watchful-waiting, receiver operating characteristic (ROC) analysis showed that area under the ROC curve was 0.686. For small-sized IPN detected by CT scan, this approach shows promise as a potential navigator to improve work-up for life-threatening cancer screening and assist SDM before surgery.







Similar content being viewed by others
References
Henschke CI, International Early Lung Cancer Action Program:screening protocol. January 15, 2023; https://www.ielcap.org/home/ielcap/clinical-resources/ielcap-protocols/. Accessed 1 Jun 2023.
The Japanese Society of CT screening: Guidelines for the management of pulmonary nodules detected by low-dose CT lung cancer screening. Ver.5 2017; https://www.jscts.org/index.php?page=guideline_index. Accessed 1 Jun 2023.
MacMahon H, Naidich DP, Goo JM, et al. Guidelines for management of incidental pulmonary nodules detected on CT images: from the Fleischner Society 2017. Radiology. 2017;284:228–43.
Ettinger DS, Wood DE, Aisner DL, Akerley W, Bauman JR, Bharat A, Bruno DS, Chang JY, Chirieac LR, DeCamp M, Dilling TJ, Dowell J, Durm GA, Gettinger S, Grotz TE, Gubens MA, Hegde A, Lackner RP, Lanuti M, Lin J, Loo BW, Lovly CM, Maldonado F, Massarelli E, Morgensztern D, Ng T, Otterson GA, Patel SP, Patil T, Polanco PM, Riely GJ, Riess J, Schild SE, Shapiro TA, Singh AP, Stevenson J, Tam A, Tanvetyanon T, Yanagawa J, Yang SC, Yau E, Gregory KM, Hughes M. 2023 NCCN Guidelines® insights: non-small cell lung cancer, Version.2. J Natl Compr Canc Netw. 2023;21:340–50.
Koh DM, Papanikolaou N, Bick U, Illing R, Kahn CE Jr, Kalpathi-Cramer J, Matos C, Martí-Bonmatí L, Miles A, Mun SK, Napel S, Rockall A, Sala E, Strickland N, Prior F. Artificial intelligence and machine learning in cancer imaging. Commun Med (Lond). 2022;27(2):133. https://doi.org/10.1038/s43856-022-00199-0.
Suzuki K, Armato SG, Li F, Sone S, Doi K. Massive training artificial neural network (MTANN) for reduction of false positives in computerized detection of lung nodules in low-dose computed tomography. Med Phys. 2003;30:1602–17.
Ardila D, Kiraly AP, Bharadwaj S, Choi B, Reicher JJ, Peng L, Tse D, Etemadi M, Ye W, Corrado G, Naidich DP, Shetty S. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med. 2019;25:954–61.
Kudo Y, Shimada Y, Matsubayashi J, Kitamura Y, Makino Y, Maehara S, Hagiwara M, Park J, Yamada T, Takeuchi S, Kakihana M, Nagao T, Ohira T, Masumoto J, Ikeda N. Artificial intelligence analysis of three-dimensional imaging data derives factors associated with postoperative recurrence in patients with radiologically solid-predominant small-sized lung cancers. Eur J Cardiothorac Surg. 2022;24(61):751–60.
Shimada Y, Kudo Y, Maehara S, Fukuta K, Masuno R, Park J, Ikeda N. Artificial intelligence-based radiomics for the prediction of nodal metastasis in early-stage lung cancer. Sci Rep. 2023;13:1028. https://doi.org/10.1038/s41598-023-28242-7. (Accessed 1 Jun).
Gao J, Qi Q, Li H, Wang Z, Sun Z, Cheng S, Yu J, Zeng Y, Hong N, Wang D, Wang H, Yang F, Li X, Li Y. 2023 Artificial-intelligence-based computed tomography histogram analysis predicting tumor invasiveness of lung adenocarcinomas manifesting as radiological part-solid nodules. Front Oncol. 2023;13:1096453. https://doi.org/10.3389/fonc.2023.1096453. (Accessed 1 Jun).
Li TZ, Xu K, Gao R, Tang Y, Lasko TA, Maldonado F, Kim S, Landman BA. Time-distance vision transformers in lung cancer diagnosis from longitudinal computed tomography. In Medical Imaging 2023: Image Processing. SPIE. 2023;12464:221–30.
Khademi S, Heidarian S, Afshar P, Naderkhani F, Oikonomou A, Plataniotis KN, Mohammadi, A. Spatio-Temporal Hybrid Fusion of CAE and SWin Transformers for Lung Cancer Malignancy Prediction. In ICASSP 2023–2023 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) (pp. 1–5). IEEE. https://arxiv.org/pdf/2210.15297.pdf. Accessed 1 Jun 2023.
Hanaoka T, Kurai M, Okada M, Ishizone S, Karasawa F, Iizuka A. Preoperative watchful-waiting time and surgical outcome of patients with non-small cell lung cancer found by chest low-dose CT screening. World J Surg. 2018;42:2164–72.
The Japan Lung Cancer Society. General rule for clinical and pathological record of lung cancer (The 8th edition, revised version). Tokyo: Kanehara-Shuppan. Co; 2021.
Hanaoka T, Nakayama J, Mukai J, Irie S, Yamanda T, Sato TA. Association of smoking with apoptosis-regulated proteins (Bcl-2, bax and p53) in resected non-small-cell lung cancers. Int J Cancer. 2001;91:267–9.
Hanaoka T, Kurai M, Okada M, Ishizone S, Karasawa F, Iizuka A, Ikeyama M, Nakayama J. Pulmonary adenocarcinoma possibly developed from the cut-end of small-sized adenocarcinoma in the lung periphery as recurrence 13 years after its wedge resection. Surg Case Rep. 2018;4:2–8.
SORABATAKE. Extract differences by synthesizing two satellite images taken at different times. 2021. https://sorabatake.jp/en/9329/. Accessed 1 Jun 2023.
Nomori H, Ohtsuka T, Naruke T, Suemasu K. Histogram analysis of computed tomography numbers of clinical T1N0M0 lung adenocarcinoma, with special reference to lymph node metastasis and tumor invasiveness. J Thorac Cardiovasc Surg. 2003;126:1584–9.
Oikonomou A, Salazar P, Zhang Y, Hwang DM, Petersen A, Dmytriw AA, Paul NS, Nguyen ET. 2019 Histogram-based models on non-thin-section chest CT predict invasiveness of primary lung adenocarcinoma subsolid nodules. Sci Rep. 2023;9:6009. https://doi.org/10.1038/s41598-019-42340-5. (Accessed 1 Jun).
OpenCV. Histogram comparison, Theory, and Code. https://docs.opencv.org/3.4/d8/dc8/tutorial_histogram_comparison.html. Accessed 1 Jun 2023.
Scikit-learn. scikit-learn developers (BSD License). https://scikit-learn.org/stable/. Accessed 1 Jun 2023.
Matplotlib. The Matplotlib development team. https://matplotlib.org/. Accessed 1 Jun 2023.
Balagurunathan Y, Schabath MB, Wang H, Liu Y, Gillies RJ. 2019 quantitative imaging features improve discrimination of malignancy in pulmonary nodules. Sci Rep. 2023;9:8528. https://doi.org/10.1038/s41598-019-44562-z. (Accessed 1 Jun).
de Margerie-Mellon C, Gill RR, Salazar P, Oikonomou A, Nguyen ET, Heidinger BH, Medina MA, VanderLaan PA, Bankier AA. 2020 Assessing invasiveness of subsolid lung adenocarcinomas with combined attenuation and geometric feature models. Sci Rep. 2023;10:14585. https://doi.org/10.1038/s41598-020-70316-3. (Accessed 1 Jun).
Wu YJ, Liu YC, Liao CY, Tang EK, Wu FZ. 2021 A comparative study to evaluate CT-based semantic and radiomic features in preoperative diagnosis of invasive pulmonary adenocarcinomas manifesting as subsolid nodules. Sci Rep. 2023;11:66. https://doi.org/10.1038/s41598-020-79690-4. (Accessed 1 Jun).
Hunter B, Chen M, Ratnakumar P, Alemu E, Logan A, Linton-Reid K, Tong D, Senthivel N, Bhamani A, Bloch S, Kemp SV, Boddy L, Jain S, Gareeboo S, Rawal B, Doran S, Navani N, Nair A, Bunce C, Kaye S, Blackledge M, Aboagye EO, Devaraj A, Lee RW. A radiomics-based decision support tool improves lung cancer diagnosis in combination with the Herder score in large lung nodules. EBioMedicine. 2023;86:104344. https://doi.org/10.1016/j.ebiom.2022.104344. (Accessed 1 Jun).
Hasegawa M, Sone S, Takashima S, Li F, Yang ZG, Maruyama Y, Watanabe T. Growth rate of small lung cancers detected on mass CT screening. Br J Radiol. 2000;73:1252–9.
Henschke CI, Yankelevitz DF, Yip R, Reeves AP, Farooqi A, Xu D, Smith JP, Libby DM, Pasmantier MW, Miettinen OS. Writing Committee for the I-ELCAP Investigators. lung cancers diagnosed at annual CT screening: volume doubling times. Radiology. 2012;263:578–83.
Sone S, Hanaoka T, Ogata H, Takayama F, Watanabe T, Haniuda M, Kaneko K, Kondo R, Yoshida K, Honda T. Small peripheral lung carcinomas with five-year post-surgical follow-up: assessment by semi-automated volumetric measurement of tumour size, CT value and growth rate on TSCT. Eur Radiol. 2012;22:104–19.
Miura K, Hamanaka K, Koizumi T, Kawakami S, Kobayashi N, Ito KI. 2019 Solid component tumor doubling time is a prognostic factor in non-small cell lung cancer patients. J Cardiothorac Surg. 2023;14:57. https://doi.org/10.1186/s13019-019-0879-x. (Accessed 1 Jun).
Setojima Y, Shimada Y, Tanaka T, Shigefuku S, Makino Y, Maehara S, Hagiwara M, Masuno R, Yamada T, Kakihana M, Kajiwara N, Ohira T, Ikeda N. Prognostic impact of solid-part tumour volume doubling time in patients with radiological part-solid or solid lung cancer. Eur J Cardiothorac Surg. 2020;57:763–70.
Tan H, Wang Y, Jiang Y, Li H, You T, Fu T, Peng J, Tan Y, Lu R, Peng B, Huang W, Xiong F. A study on the differential of solid lung adenocarcinoma and tuberculous granuloma nodules in CT images by Radiomics machine learning. Sci Rep. 2023;13:5853. https://doi.org/10.1038/s41598-023-32979-6. (Accessed 1 Jun).
Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R, Rosenthal R, Salm M, Horswell S, Escudero M, Matthews N, Rowan A, Chambers T, Moore DA, Turajlic S, Xu H, Lee SM, Forster MD, Ahmad T, Hiley CT, Abbosh C, Falzon M, Borg E, Marafioti T, Lawrence D, Hayward M, Kolvekar S, Panagiotopoulos N, Janes SM, Thakrar R, Ahmed A, Blackhall F, Summers Y, Shah R, Joseph L, Quinn AM, Crosbie PA, Naidu B, Middleton G, Langman G, Trotter S, Nicolson M, Remmen H, Kerr K, Chetty M, Gomersall L, Fennell DA, Nakas A, Rathinam S, Anand G, Khan S, Russell P, Ezhil V, Ismail B, Irvin-Sellers M, Prakash V, Lester JF, Kornaszewska M, Attanoos R, Adams H, Davies H, Dentro S, Taniere P, O’Sullivan B, Lowe HL, Hartley JA, Iles N, Bell H, Ngai Y, Shaw JA, Herrero J, Szallasi Z, Schwarz RF, Stewart A, Quezada SA, Le Quesne J, Van Loo P, Dive C, Hackshaw A, Swanton C, TRACERx Consortium. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376:2109–21.
Yankelevitz DF, Henschke CI. Overdiagnosis in lung cancer screening. Transl Lung Cancer Res. 2021;10:1136–40.
Drukker K, Chen W, Gichoya J, Gruszauskas N, Kalpathy-Cramer J, Koyejo S, Myers K, Sá RC, Sahiner B, Whitney H, Zhang Z, Giger M. Toward fairness in artificial intelligence for medical image analysis: identification and mitigation of potential biases in the roadmap from data collection to model deployment. J Med Imaging (Bellingham). 2023;10(061104):2023. https://doi.org/10.1117/1.JMI.10.6.061104.Accessed1Jun.
MIDRC WEB. Medical imaging AI/machine learning bias awareness tool: An interactive decision tree. https://www.midrc.org/bias-awareness-tool. Accessed 1 Jun 2023.
Heidarian S, Afshar P, Oikonomou A, Plataniotis KN, Mohammadi A. CAE-Transformer: Transformer-based model to predict invasiveness of lung adenocarcinoma subsolid nodules from non-thin section 3D CT scans. 2021. https://doi.org/10.48550/arXiv.2110.08721. Accessed 1 Jun 2023.
Acknowledgements
We would like to express our sincere gratitude to Mr. Noriyasu Kobayashi and Ms. Haruka Takemura, Department of Laboratory Medicine, JA North Alps Medical Center Azumi Hospital, Japan, for the preparation of the pathological specimen. This work was facilitated by the open source contributions of Python and OpenCV [20].
Author information
Authors and Affiliations
Contributions
All authors meet the ICMJE authorship criteria. TH: Conceptualization, Data Curation, Formal Analysis, Investigation, Methodology, Project Administration, Resources, Supervision, Validation, Visualization, Writing-original draft, Writing-review and editing. HM: Investigation, Data Curation, Formal Analysis, Methodology, Resources, Validation, Writing-review and editing. JN: Investigation, Data Curation, Formal Analysis, Methodology, Resources, Supervision, Validation, Writing-review and editing. SO: Validation, Writing—review and editing. KI: Validation, Writing—review and editing. MO: Validation, Writing—review and editing.
Corresponding author
Ethics declarations
Conflict of interest
All authors have no financial/commercial conflicts of interest.
Ethical approval
This investigation was approved by the institutional ethics committees (No. 29–06-01), with waiver of patient informed consent.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
About this article
Cite this article
Hanaoka, T., Matoba, H., Nakayama, J. et al. A spatio-temporal image analysis for growth of indeterminate pulmonary nodules detected by CT scan. Radiol Phys Technol 17, 71–82 (2024). https://doi.org/10.1007/s12194-023-00750-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12194-023-00750-1